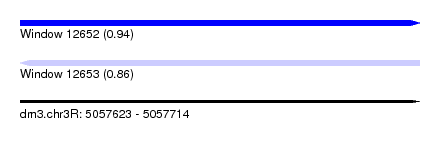
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,057,623 – 5,057,714 |
| Length | 91 |
| Max. P | 0.942181 |

| Location | 5,057,623 – 5,057,714 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.40826 |
| G+C content | 0.36772 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -9.56 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5057623 91 + 27905053 CCGAGCCUUAUCAAUUGAAUUAAA--GACUACUACUUAUGCUACUGAUAAGUGGUAGUAUUCCUAAGCAAAUCAAUUGCAACAUAUUCCCCUC----------------------- ..(((...................--.((((((((((((.(....)))))))))))))........((((.....))))...........)))----------------------- ( -18.00, z-score = -2.67, R) >droEre2.scaffold_4770 1203452 113 - 17746568 CCGAGCGUUAUCAAUUGAAUUAAA--GAAUACUACUCAUGCUACUGAUAAGUAA-AGUAUUCCUAAACAAAUCAAUUGCAACAUAUCCCACAUGUGUAUGUGCGUUCCCUCCCUUC ..((((((...(((((((......--(((((((.(....).((((....)))).-))))))).........)))))))..(((((..(.....)..)))))))))))......... ( -19.46, z-score = -2.30, R) >droYak2.chr3R 9109434 115 + 28832112 CCGGGCCUUAUCAAUUGAAUUAAAAUGAAUACUACUCGUGCUACUGAUAAGUAA-AGUAUUCCCAACCAAAUCAAUUGCAACAUAUCGCAGAUAUGUGUAUGCCCUCUCCCUUUAA ..((((.....(((((((........(((((((.....((((.......)))).-))))))).........)))))))..(((((((...)))))))....))))........... ( -24.33, z-score = -3.11, R) >droSim1.chr3R 11264259 91 - 27517382 CCCAGCCUUAUCAAUUGAAUUAAA--GAAUACUACUCAUGCUUCUGAUAAGUGAUAGUAUCCCUAACCAAAUCAAUUGCAACAUAUUCCCCUC----------------------- ...........(((((((.....(--(.((((((.((((.(....)....))))))))))..)).......)))))))...............----------------------- ( -10.30, z-score = -1.31, R) >consensus CCGAGCCUUAUCAAUUGAAUUAAA__GAAUACUACUCAUGCUACUGAUAAGUAA_AGUAUUCCUAACCAAAUCAAUUGCAACAUAUCCCACAC_______________________ ...........(((((((........((((((((....((((.......)))).)))))))).........)))))))...................................... ( -9.56 = -10.30 + 0.75)

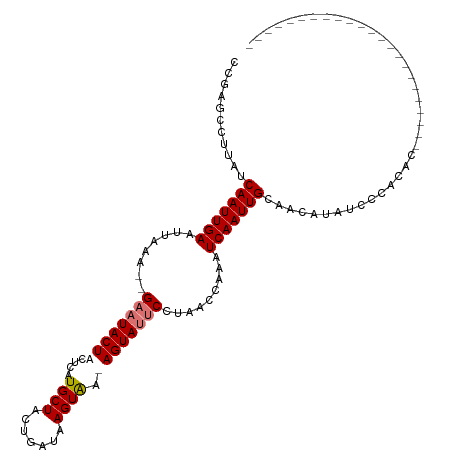
| Location | 5,057,623 – 5,057,714 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.40826 |
| G+C content | 0.36772 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5057623 91 - 27905053 -----------------------GAGGGGAAUAUGUUGCAAUUGAUUUGCUUAGGAAUACUACCACUUAUCAGUAGCAUAAGUAGUAGUC--UUUAAUUCAAUUGAUAAGGCUCGG -----------------------...(((.........(((((((.(((...((((...((((.(((((((....).)))))).))))))--))))).)))))))......))).. ( -17.06, z-score = 0.05, R) >droEre2.scaffold_4770 1203452 113 + 17746568 GAAGGGAGGGAACGCACAUACACAUGUGGGAUAUGUUGCAAUUGAUUUGUUUAGGAAUACU-UUACUUAUCAGUAGCAUGAGUAGUAUUC--UUUAAUUCAAUUGAUAACGCUCGG .....(((....(.(((((....))))).)....(((((((((((.(((...(((((((((-..(((((((....).)))))))))))))--))))).))))))).)))).))).. ( -30.20, z-score = -3.18, R) >droYak2.chr3R 9109434 115 - 28832112 UUAAAGGGAGAGGGCAUACACAUAUCUGCGAUAUGUUGCAAUUGAUUUGGUUGGGAAUACU-UUACUUAUCAGUAGCACGAGUAGUAUUCAUUUUAAUUCAAUUGAUAAGGCCCGG ...........((((....(((((((...)))))))..(((((((.((((....(((((((-..((((...........)))))))))))...)))).))))))).....)))).. ( -29.30, z-score = -2.34, R) >droSim1.chr3R 11264259 91 + 27517382 -----------------------GAGGGGAAUAUGUUGCAAUUGAUUUGGUUAGGGAUACUAUCACUUAUCAGAAGCAUGAGUAGUAUUC--UUUAAUUCAAUUGAUAAGGCUGGG -----------------------...........(((.(((((((.(((...(((((((((...(((((((....).)))))))))))))--))))).)))))))....))).... ( -18.60, z-score = -0.80, R) >consensus _______________________AAGGGGAAUAUGUUGCAAUUGAUUUGGUUAGGAAUACU_UCACUUAUCAGUAGCAUGAGUAGUAUUC__UUUAAUUCAAUUGAUAAGGCUCGG ..................................(((.(((((((.((((....(((((((...(((((((....).)))))))))))))...)))).)))))))....))).... (-16.00 = -16.88 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:22 2011