
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,049,796 – 5,049,949 |
| Length | 153 |
| Max. P | 0.725022 |

| Location | 5,049,796 – 5,049,949 |
|---|---|
| Length | 153 |
| Sequences | 6 |
| Columns | 174 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.43495 |
| G+C content | 0.41108 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -19.98 |
| Energy contribution | -18.52 |
| Covariance contribution | -1.46 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
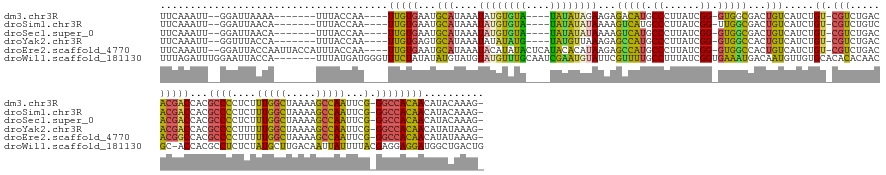
>dm3.chr3R 5049796 153 + 27905053 UUCAAAUU--GGAUUAAAA-------UUUACCAA----UUGUGAAUGCAUAAAUAUGUGUA----UAUAUAGAAGAGACAUGCCCUUAUCGG-GUGGCGACUGUCAUCUGU-CGUCUGACACGACCACGCCCCUCUUUGGCUAAAAGCCAAUUCG-GGCCACAACAUACAAAG- ....((((--((.......-------....))))----))(((.(((((((....))))))----).......(((((((((((((.....)-).))))..)))).)))((-(((.....))))))))((((....(((((.....)))))...)-)))..............- ( -39.60, z-score = -1.83, R) >droSim1.chr3R 11256409 153 - 27517382 UUCAAAUU--GGAUUAACA-------UUUACCAA----UUGUGAAUGCAUAAAUAUGUGUA----UAUAUAUAAAAGUCAUGCCCUUAUCGG-UUGGCGACUGUCAUCUGU-CGUCUGUCACGACCACGCCCCUCUUUGGCUAAAAGCCAAUUCG-GGCCACAACAUACAAAG- ....((((--((.......-------....))))----))(((.(((((((....))))))----)..........(((..(((......))-).((((((........))-))))......))))))((((....(((((.....)))))...)-)))..............- ( -37.30, z-score = -1.92, R) >droSec1.super_0 4288118 153 - 21120651 UUCAAAUU--GGAUUAACA-------UUUACCAA----UUGUGAAUGCAUAAAUAUGUGUA----UAUAUAUAAAAGUCAUGCCCUUAUCGG-GUGGCGACUGUCAUCUGU-CGUCUGACACGACCACGCCCCUCUUUGGCUAAAAGCCAAUUCG-GGCCACAACAUACAAAG- ....((((--((.......-------....))))----))(((.(((((((....))))))----)..........((((.((((.....))-))((((((........))-)))))))).....)))((((....(((((.....)))))...)-)))..............- ( -42.70, z-score = -3.00, R) >droYak2.chr3R 9101423 153 + 28832112 UUCAAAUU--GGUUUACCA-------UUUACCAA----UUGUGAGUGCAUAAAUAUAUAUG----UAUGUUAAAGAGCCAUGCCCUUAUCGG-GUGGCCACUGUCAUCUGU-CGUCUGACACGACCACGCCCCUUUUUGGCUAAAAGCCAAUUCG-GGCCACAACAUAUAAAG- ((((((((--(((......-------...)))))----)).))))..............((----((((((...(.(((..((((.....))-))))))..........((-(((.....)))))...((((....(((((.....)))))...)-)))...))))))))...- ( -41.70, z-score = -2.08, R) >droEre2.scaffold_4770 1195423 164 - 17746568 UUCAAAUU--GGAUUACCAAUUACCAUUUACCAA----UUGUGAAUGCAUAAAUACAUAUACUCAUACACAUAAGAGCCAUGCCCUUAUCGG-GUGGCCACUGUCAUCUGU-CGUCUGACACGGCCACGCCCCUUUUUGGCUAAAAGCCAAUUCG-GGCCACAACAUAUAAAG- ....((((--((....))))))........((..----.((((.(((..((........))..))).))))((((.((...)).))))..))-((((((..(((((.....-....))))).))))))((((....(((((.....)))))...)-)))..............- ( -41.90, z-score = -2.43, R) >droWil1.scaffold_181130 8430615 166 - 16660200 UUUAGAUUUGGAAUUACCA-------UUUAUGAUGGGUUUCUAUAUAUGUAUGUAUGUUUGCAAUCGAAUGUAUUCGUUUUGCCUUUAUCGGUGAAAUGACAAUGUUGUGCACACACAACGC-ACCACGCCUCUCUAUGCUUGACAAUUAUUUUACGAGGAGGAUGGCUGACUG ........(((((...(((-------((...)))))..)))))....(((..(((((((((....)))))))))((((((..((......))..))))))....((((((....))))))))-).((.(((..(((...((((............)))).)))..))))).... ( -36.10, z-score = 0.21, R) >consensus UUCAAAUU__GGAUUAACA_______UUUACCAA____UUGUGAAUGCAUAAAUAUGUGUA____UAUAUAUAAGAGUCAUGCCCUUAUCGG_GUGGCGACUGUCAUCUGU_CGUCUGACACGACCACGCCCCUCUUUGGCUAAAAGCCAAUUCG_GGCCACAACAUACAAAG_ ......................................(((((...(((....((((((((....))))))))...(((((.((......)).)))))...)))........................(((.....(((((.....))))).....)))))))).......... (-19.98 = -18.52 + -1.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:21 2011