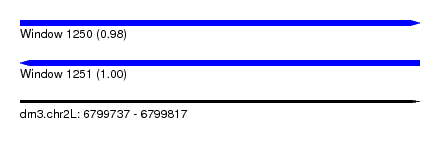
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,799,737 – 6,799,817 |
| Length | 80 |
| Max. P | 0.998274 |

| Location | 6,799,737 – 6,799,817 |
|---|---|
| Length | 80 |
| Sequences | 13 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.61627 |
| G+C content | 0.56129 |
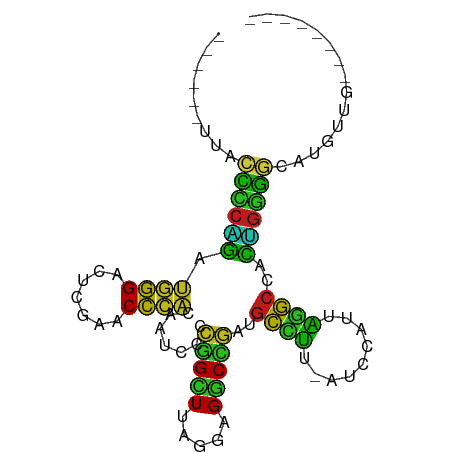
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -28.15 |
| Energy contribution | -24.99 |
| Covariance contribution | -3.16 |
| Combinations/Pair | 2.43 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
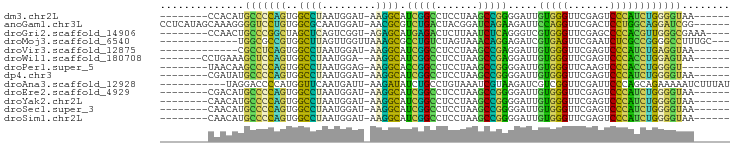
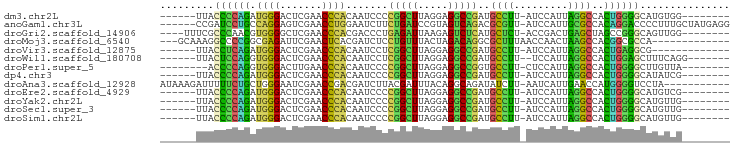
>dm3.chr2L 6799737 80 + 23011544 --------CCACAUGCCCCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA------ --------.....((((((((..((((.......-.))))..((((.......))))((((((.((....))))))))..)))))))).------ ( -32.10, z-score = -0.77, R) >anoGam1.chr3L 36924764 88 - 41284009 CCUCAUAGCAAAGGGGUCCUGUGGCGCAAUGGAU-AACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGG------ (((........)))(((((((((((((.......-...))))).......(((((....)))))((((.......))))))))))))..------ ( -27.90, z-score = -0.55, R) >droGri2.scaffold_14906 200108 82 - 14172833 --------CCAACUGCCCGGCUAGCUCAGUCGGU-AGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA---- --------.....((((((((..((((.......-.))))....((((((.......))))))(((((.......)))))))))))))...---- ( -32.80, z-score = -1.91, R) >droMoj3.scaffold_6540 19666616 79 - 34148556 -------------UGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUUUGC--- -------------.((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))....--- ( -24.70, z-score = -0.04, R) >droVir3.scaffold_12875 10427360 75 + 20611582 -------------CGCCUCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGAGGAUUGUGGGUUCGAGUCCCAUCUGAGGUAA------ -------------.(((((((..((((.......-.)))).(((((.......))))).....(((((.......))))))))))))..------ ( -28.80, z-score = -1.74, R) >droWil1.scaffold_180708 7702623 80 + 12563649 -------CCUGAAAGCUCCAGUGGCCUAAUGGA--AAGGCAUCGGCCUCCUAAGCCGAGGAUUGUGGGUUCGAGUCCCACCUGGAGUAA------ -------.......(((((((..((((......--.)))).(((((.......))))).....(((((.......))))))))))))..------ ( -30.50, z-score = -1.85, R) >droPer1.super_5 4707410 78 + 6813705 --------UAACAAGCCCCAGUGGCCUAAUGGAG-AAGGCACCGGCCUCCUAAGCCGGGGAUUGUGGGUUCAAGUCCCACCUGGGGU-------- --------......(((((((..((((.......-.)))).(((((.......))))).....(((((.......))))))))))))-------- ( -34.30, z-score = -1.39, R) >dp4.chr3 13014949 80 + 19779522 --------CGAUAUGCCCCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA------ --------.....((((((((..((((.......-.))))..((((.......))))((((((.((....))))))))..)))))))).------ ( -32.10, z-score = -1.06, R) >droAna3.scaffold_12928 260993 83 + 771024 -----------UAGGACCCCAUGGUUCAAUGAUU-AAGAUAUCUGCCUGUAAAUCGUAAGAUCGUCGGUUCGAUUCCCAGCAGAAAAAUCUUUAU -----------..(((((....))))).......-(((((.(((((.((..(((((...(.....)....)))))..)))))))...)))))... ( -20.00, z-score = -2.08, R) >droEre2.scaffold_4929 15715912 80 + 26641161 --------CGACAUGCCCCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA------ --------.....((((((((..((((.......-.))))..((((.......))))((((((.((....))))))))..)))))))).------ ( -32.10, z-score = -0.95, R) >droYak2.chr2L 16215580 80 - 22324452 --------CAACAUGCCCCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA------ --------.....((((((((..((((.......-.))))..((((.......))))((((((.((....))))))))..)))))))).------ ( -32.10, z-score = -1.10, R) >droSec1.super_3 2338511 80 + 7220098 --------CAACAUGCCCCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA------ --------.....((((((((..((((.......-.))))..((((.......))))((((((.((....))))))))..)))))))).------ ( -32.10, z-score = -1.10, R) >droSim1.chr2L 6601146 80 + 22036055 --------CAACAUGCCCCAGUGGCCUAAUGGAU-AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA------ --------.....((((((((..((((.......-.))))..((((.......))))((((((.((....))))))))..)))))))).------ ( -32.10, z-score = -1.10, R) >consensus ________CAACAUGCCCCAGUGGCCUAAUGGAU_AAGGCAUCGGCCUCCUAAGCCGGGGAUUGUGGGUUCGAGUCCCAUCUGGGGUAA______ ..............(((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))))........ (-28.15 = -24.99 + -3.16)
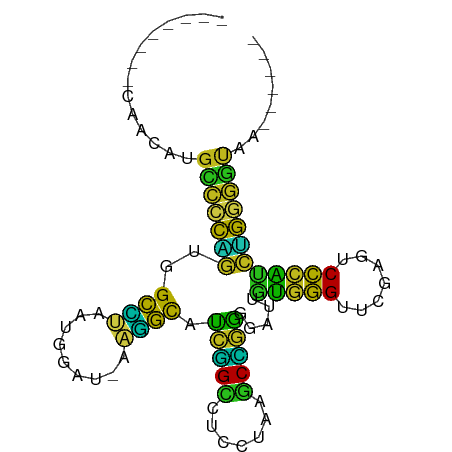
| Location | 6,799,737 – 6,799,817 |
|---|---|
| Length | 80 |
| Sequences | 13 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.61627 |
| G+C content | 0.56129 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -23.46 |
| Energy contribution | -21.27 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 2.21 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6799737 80 - 23011544 ------UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGGGGCAUGUGG-------- ------....(((....)))........(((((..((((((((((((((((....)))))-.....)))))))...))))..)))))-------- ( -30.00, z-score = -1.35, R) >anoGam1.chr3L 36924764 88 + 41284009 ------CCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUU-AUCCAUUGCGCCACAGGACCCCUUUGCUAUGAGG ------....((((((((((.......))))).....((((((.....)))))).((((.-.......))))..)))))..((((......)))) ( -25.60, z-score = -1.13, R) >droGri2.scaffold_14906 200108 82 + 14172833 ----UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCU-ACCGACUGAGCUAGCCGGGCAGUUGG-------- ----......((((((((((.......)))))(.(((((((((.....)))))).((((.-.......)))).....)))).)))))-------- ( -31.70, z-score = -2.54, R) >droMoj3.scaffold_6540 19666616 79 + 34148556 ---GCAAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA------------- ---.....(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).------------- ( -25.20, z-score = -2.53, R) >droVir3.scaffold_12875 10427360 75 - 20611582 ------UUACCUCAGAUGGGACUCGAACCCACAAUCCUCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGAGGCG------------- ------...((((((.((((.......))))........((((((((((((....)))))-.....))))))).))))))..------------- ( -27.70, z-score = -2.79, R) >droWil1.scaffold_180708 7702623 80 - 12563649 ------UUACUCCAGGUGGGACUCGAACCCACAAUCCUCGGCUUAGGAGGCCGAUGCCUU--UCCAUUAGGCCACUGGAGCUUUCAGG------- ------...(((((((((((.......))))).......((((((((((((....)))))--....))))))).))))))........------- ( -31.90, z-score = -2.74, R) >droPer1.super_5 4707410 78 - 6813705 --------ACCCCAGGUGGGACUUGAACCCACAAUCCCCGGCUUAGGAGGCCGGUGCCUU-CUCCAUUAGGCCACUGGGGCUUGUUA-------- --------.(((((((((((.......))))).......((((((((((((....)))))-)).....))))).)))))).......-------- ( -35.30, z-score = -1.87, R) >dp4.chr3 13014949 80 - 19779522 ------UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGGGGCAUAUCG-------- ------...((((((.((((.......))))........((((((((((((....)))))-.....))))))).)))))).......-------- ( -29.80, z-score = -2.23, R) >droAna3.scaffold_12928 260993 83 - 771024 AUAAAGAUUUUUCUGCUGGGAAUCGAACCGACGAUCUUACGAUUUACAGGCAGAUAUCUU-AAUCAUUGAACCAUGGGGUCCUA----------- ...(((((...((((((..((((((..............))))))...)))))).)))))-.......(.(((....))).)..----------- ( -18.64, z-score = -0.99, R) >droEre2.scaffold_4929 15715912 80 - 26641161 ------UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGGGGCAUGUCG-------- ------...((((((.((((.......))))........((((((((((((....)))))-.....))))))).)))))).......-------- ( -29.80, z-score = -1.84, R) >droYak2.chr2L 16215580 80 + 22324452 ------UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGGGGCAUGUUG-------- ------...((((((.((((.......))))........((((((((((((....)))))-.....))))))).)))))).......-------- ( -29.80, z-score = -1.87, R) >droSec1.super_3 2338511 80 - 7220098 ------UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGGGGCAUGUUG-------- ------...((((((.((((.......))))........((((((((((((....)))))-.....))))))).)))))).......-------- ( -29.80, z-score = -1.87, R) >droSim1.chr2L 6601146 80 - 22036055 ------UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU-AUCCAUUAGGCCACUGGGGCAUGUUG-------- ------...((((((.((((.......))))........((((((((((((....)))))-.....))))))).)))))).......-------- ( -29.80, z-score = -1.87, R) >consensus ______UUACCCCAGAUGGGACUCGAACCCACAAUCCCCGGCUUAGGAGGCCGAUGCCUU_AUCCAUUAGGCCACUGGGGCAUGUUG________ .........((((((.((((.......)))).......(((((.....)))))..((((.........))))..))))))............... (-23.46 = -21.27 + -2.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:06 2011