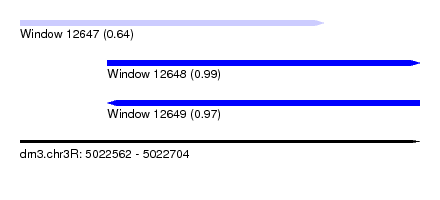
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,022,562 – 5,022,704 |
| Length | 142 |
| Max. P | 0.986440 |

| Location | 5,022,562 – 5,022,670 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.08 |
| Shannon entropy | 0.67306 |
| G+C content | 0.40541 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -8.14 |
| Energy contribution | -9.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
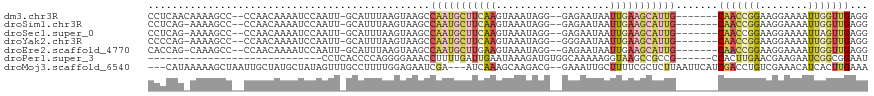
>dm3.chr3R 5022562 108 + 27905053 CCUCAACAAAAGCC--CCAACAAAAUCCAAUU-GCAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGG--GAGAAUAAUUGAAGCAUUG-------CAACCGGAAGGAAAAUUGGUUGAGG (((((((.......--..............((-((......))))((.((((((((((.........--........)))))))))))-------)..((....)).......))))))) ( -28.83, z-score = -2.96, R) >droSim1.chr3R 11228120 107 - 27517382 CCUCAG-AAAAGCC--CCAACAAAAUCCAAUU-GCAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGG--GAGAAUAAUUGAAGCAUUG-------CAACCGGAAGGAAAAUUGGUUGAGG ((((((-....(..--....).....((((((-............((.((((((((((.........--........)))))))))))-------)..((....))..)))))))))))) ( -26.93, z-score = -2.28, R) >droSec1.super_0 4261151 107 - 21120651 CCUCAG-AAAAGCC--CCAACAAAAUCCAAUU-GCAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGG--GAGAAUAAUUGAAGCAUUG-------CAACCGGAAGGAAAAUUAGUUGAGG ((((((-.......--..............((-((......))))((.((((((((((.........--........)))))))))))-------)..((....))........)))))) ( -25.13, z-score = -2.42, R) >droYak2.chr3R 9071828 107 + 28832112 CCCCAG-AAAAGCC--CCAACAAAAUCCAAUU-GCAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGG--GGGAAUAAUUGAAGCAUUG-------CAACCGGAAGGAAAAUUGGUUGAGG ......-.....((--.((((.........((-((......))))((.((((((((((.........--........)))))))))))-------)..((....)).......)))).)) ( -24.23, z-score = -0.57, R) >droEre2.scaffold_4770 1167699 107 - 17746568 CACCAG-CAAAGCC--CCAACAAAAUCCAAUU-GCAUUUAAGUAAGCCAAUGCUUGAAGUAAAUAGG--GAGAAUAAUUGAAGCAUUG-------CAACCGGAAGGAAAAUUGGUUGAGG .(((((-(((.((.--.(((.....(((..((-((.((((((((......))))))))))))....)--))......)))..)).)))-------)..((....)).....))))..... ( -27.50, z-score = -2.39, R) >droPer1.super_3 310934 85 - 7375914 -----------------------------CCUCACCCCAGGGGAAACCUUUUGAUUGAAUAAAGAUGUGGCAAAAAGGUAAGCCGCCG------CCACUUGAACGAAGAAUCGGCGGAAU -----------------------------.....((((((((....))))..((((..........(((((.....((....))...)------))))..........)))))).))... ( -22.35, z-score = -0.80, R) >droMoj3.scaffold_6540 23920147 112 + 34148556 ---CAUAAAAAGCUAAUUGCUAUGCUAUAGUUUGCCUUUUGGAGAAUCGA---AUCAAAGCAAGACG--GAAAUUGCUUUUCGCUCUUAAUUCAUCGACCUGUCGAAACAUCACUUGAAA ---..(((((.((.....(((((...)))))..)).)))))((((..(((---(...((((((....--....)))))))))).))))......((((....)))).............. ( -17.90, z-score = 0.70, R) >consensus CCUCAG_AAAAGCC__CCAACAAAAUCCAAUU_GCAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGG__GAGAAUAAUUGAAGCAUUG_______CAACCGGAAGGAAAAUUGGUUGAGG ....................................(((((.(((..(((((((((((...................)))))))))))..........((....))....))).))))). ( -8.14 = -9.70 + 1.56)
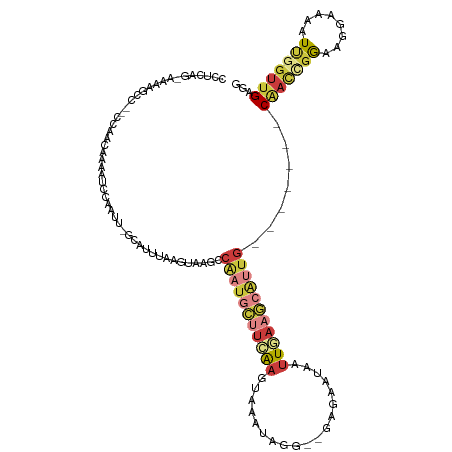
| Location | 5,022,593 – 5,022,704 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.64178 |
| G+C content | 0.37175 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -8.54 |
| Energy contribution | -10.30 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5022593 111 + 27905053 CAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGGGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAA-AAUUGGUUGAG-GCGAAGCUAAAAUGGCAUUUUAAAC-----GAUAGUUUUC (((((.(((..(((((((((((((.................)))))))))).(((((((......-..))))))).)-))...))).))))).......((((-----....)))).. ( -28.03, z-score = -3.04, R) >droSim1.chr3R 11228150 111 - 27517382 CAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGGGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAA-AAUUGGUUGAG-GCGAAGCUAAAAUGGCAUUUUAAAC-----GAAAGUUUUC (((((.(((..(((((((((((((.................)))))))))).(((((((......-..))))))).)-))...))).))))).......((((-----....)))).. ( -31.03, z-score = -4.10, R) >droSec1.super_0 4261181 111 - 21120651 CAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGGGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAA-AAUUAGUUGAG-GCGAAGCUAAAAUGGCAUUUUAAAC-----GAAAGUUUUC (((((.(((..(((((((((((((.................))))))))))....((....))..-..........)-))...))).))))).......((((-----....)))).. ( -29.63, z-score = -3.90, R) >droYak2.chr3R 9071858 111 + 28832112 CAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGGGGGAAUAAUUGAAGCAUUGCAACCGGAAGGAA-AAUUGGUUGAG-GCGAGGCUAAAAUGGCAUUUUAAAC-----GAAAGUUUUC (((((.(((..(((((((((((((.................)))))))))).(((((((......-..))))))).)-))...))).))))).......((((-----....)))).. ( -32.13, z-score = -4.21, R) >droEre2.scaffold_4770 1167729 111 - 17746568 CAUUUAAGUAAGCCAAUGCUUGAAGUAAAUAGGGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAA-AAUUGGUUGAG-GCGAGGCUAAAAUGGCAUUUUAAAC-----GAAAGUUUUC (((((.(((..((((((((((.((.................)).))))))).(((((((......-..))))))).)-))...))).))))).......((((-----....)))).. ( -28.03, z-score = -3.11, R) >droPer1.super_3 310958 96 - 7375914 -----------------AUUGAAUAAAGAUGUGGCAAAAAGGUAAGCCGCCGCCACUUGAACGAAGAAUCGGCGGAAUACAAUCGGGAAAUGCCAUUUAAAAC-----GAAACUUUUU -----------------.............((((((.....(((..((((((...(((.....)))...))))))..)))..........)))))).......-----.......... ( -20.76, z-score = -0.81, R) >droMoj3.scaffold_6540 23920178 109 + 34148556 ------CCUUUUGGAGAAUCGAAUCAAAGCAAGACGGAAA--UUGCUUUUCGCU-CUUAAUUCAUCGACCUGUCGAAACAUCACUUGAAAUGUUACCAAAAAUCAGUUGACACAUUUC ------..(((((((((..((((...((((((........--)))))))))).)-)).......((((....))))(((((........))))).))))))................. ( -18.10, z-score = 0.24, R) >consensus CAUUUAAGUAAGCCAAUGCUUCAAGUAAAUAGGGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAA_AAUUGGUUGAG_GCGAAGCUAAAAUGGCAUUUUAAAC_____GAAAGUUUUC ...........(((((((((((((.................))))))))).....((....)).....((((((........))))))..))))........................ ( -8.54 = -10.30 + 1.76)
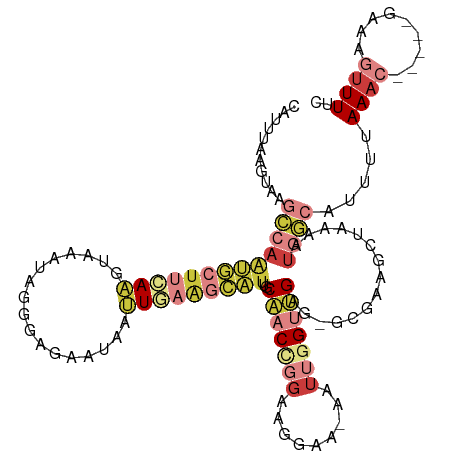
| Location | 5,022,593 – 5,022,704 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.64178 |
| G+C content | 0.37175 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -6.41 |
| Energy contribution | -8.09 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5022593 111 - 27905053 GAAAACUAUC-----GUUUAAAAUGCCAUUUUAGCUUCGC-CUCAACCAAUU-UUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCCCUAUUUACUUGAAGCAUUGGCUUACUUAAAUG .........(-----((((((...(((.............-..(((((....-........))))).((((((((((.((...........)).)))))))))))))....))))))) ( -22.30, z-score = -3.80, R) >droSim1.chr3R 11228150 111 + 27517382 GAAAACUUUC-----GUUUAAAAUGCCAUUUUAGCUUCGC-CUCAACCAAUU-UUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCCCUAUUUACUUGAAGCAUUGGCUUACUUAAAUG .........(-----((((((...(((.............-..(((((....-........))))).((((((((((.((...........)).)))))))))))))....))))))) ( -22.30, z-score = -3.75, R) >droSec1.super_0 4261181 111 + 21120651 GAAAACUUUC-----GUUUAAAAUGCCAUUUUAGCUUCGC-CUCAACUAAUU-UUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCCCUAUUUACUUGAAGCAUUGGCUUACUUAAAUG .........(-----((((((...(((......((...((-(..........-........))).))((((((((((.((...........)).)))))))))))))....))))))) ( -21.17, z-score = -3.32, R) >droYak2.chr3R 9071858 111 - 28832112 GAAAACUUUC-----GUUUAAAAUGCCAUUUUAGCCUCGC-CUCAACCAAUU-UUCCUUCCGGUUGCAAUGCUUCAAUUAUUCCCCCUAUUUACUUGAAGCAUUGGCUUACUUAAAUG .........(-----((((((...(((.............-..(((((....-........))))).((((((((((.((...........)).)))))))))))))....))))))) ( -22.30, z-score = -3.82, R) >droEre2.scaffold_4770 1167729 111 + 17746568 GAAAACUUUC-----GUUUAAAAUGCCAUUUUAGCCUCGC-CUCAACCAAUU-UUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCCCUAUUUACUUCAAGCAUUGGCUUACUUAAAUG .........(-----((((((...(((.............-..(((((....-........))))).(((((((.((.((...........)).)).))))))))))....))))))) ( -16.10, z-score = -1.94, R) >droPer1.super_3 310958 96 + 7375914 AAAAAGUUUC-----GUUUUAAAUGGCAUUUCCCGAUUGUAUUCCGCCGAUUCUUCGUUCAAGUGGCGGCGGCUUACCUUUUUGCCACAUCUUUAUUCAAU----------------- ..........-----........(((((......((..(((..((((((...(((.....)))...))))))..)))..)).)))))..............----------------- ( -20.90, z-score = -1.37, R) >droMoj3.scaffold_6540 23920178 109 - 34148556 GAAAUGUGUCAACUGAUUUUUGGUAACAUUUCAAGUGAUGUUUCGACAGGUCGAUGAAUUAAG-AGCGAAAAGCAA--UUUCCGUCUUGCUUUGAUUCGAUUCUCCAAAAGG------ (((((((....((..(...)..)).)))))))...((((...((((....))))...))))((-((((((((((((--........))))))...)))).))))........------ ( -21.90, z-score = 0.44, R) >consensus GAAAACUUUC_____GUUUAAAAUGCCAUUUUAGCUUCGC_CUCAACCAAUU_UUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCCCUAUUUACUUGAAGCAUUGGCUUACUUAAAUG ...........................................(((((.............)))))(((((((((((.................)))))))))))............. ( -6.41 = -8.09 + 1.68)
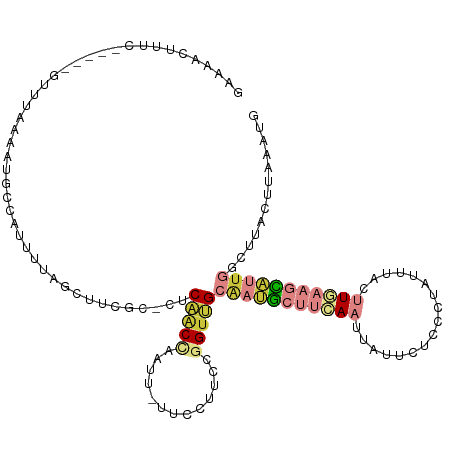
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:19 2011