| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,014,246 – 5,014,390 |
| Length | 144 |
| Max. P | 0.954479 |

| Location | 5,014,246 – 5,014,390 |
|---|---|
| Length | 144 |
| Sequences | 8 |
| Columns | 163 |
| Reading direction | forward |
| Mean pairwise identity | 60.50 |
| Shannon entropy | 0.78227 |
| G+C content | 0.36093 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -9.68 |
| Energy contribution | -9.58 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
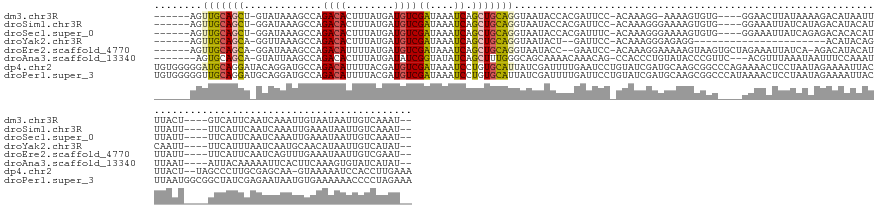
>dm3.chr3R 5014246 144 + 27905053 ------AGUUGCAGCU-GUAUAAAGCCAGACACUUUAUGAUGUCGAUAAAUCAGCUGCAGGUAAUACCACGAUUCC-ACAAAGG-AAAAGUGUG----GGAACUUAUAAAAGACAUAAUUUUACU----GUCAUUCAAUCAAAUUGUAAUAAUUGUCAAAU-- ------(.((((((((-(..........(((((.....).)))).......))))))))).)....(((((.((((-.....))-))...))))----)............((((.....((((.----................))))....))))....-- ( -32.56, z-score = -1.96, R) >droSim1.chr3R 11219686 145 - 27517382 ------AGUUGCAGCU-GGAUAAAGCCAGACACUUUAUGAUGUCGAUAAAUCAGCUGCAGGUAAUACCACGAUUCC-ACAAAGGGAAAAGUGUG----GGAAAUUAUCAUAGACAUACAUUUAUU----UUCAUUCAAUCAAAUUGAAAUAAUUGUCAAAU-- ------...(((((((-((.........(((((.....).))))......)))))))))((((((.(((((.((((-......))))...))))----)...))))))...((((.....(((((----((.(((......))).))))))).))))....-- ( -37.26, z-score = -3.23, R) >droSec1.super_0 4252801 145 - 21120651 ------AGUUGCAGCU-GGAUAAAGCCAGACACUUUAUGAUGUCGAUAAAUCAGCUGCAGGUAAUACCACGAUUUC-ACAAAGGGAAAAGUGUG----GGAAAUUAUCAGAGACACACAUUUAUU----UUCAUUCAAUCAAAUUGAAAUAAUUGUCAAAU-- ------...(((((((-((.........(((((.....).))))......)))))))))((((((.(((((.((((-......))))...))))----)...))))))...((((.....(((((----((.(((......))).))))))).))))....-- ( -33.66, z-score = -2.10, R) >droYak2.chr3R 9063382 125 + 28832112 ------AGUUGCAGCA-GGUUAAAGCCAGACACUUUAUGAUGUCGAUAAAUCAGCUGCAGGUAAUACU--GAUUCC-ACAAAGGGAGAGG----------------------ACAUACAGCAAUU----UUCAUUUAAUCAAUGCAACAUAAUUGUCAUAU-- ------.((((((...-(((((((....(((((.....).)))).........(((((((......))--).((((-......))))...----------------------.....))))....----....)))))))..)))))).............-- ( -28.20, z-score = -1.71, R) >droEre2.scaffold_4770 1159364 146 - 17746568 ------AGUUGCAGCA-GGAUAAAGCCAGACAUUUUAUGAUGUCGAUAAAUCAGCUGCAGGUAAUACC--GAAUCC-ACAAAGGAAAAAGUAAGUGCUAGAAAUUAUCA-AGACAUACAUUUAUU----UUCAUUCAAUCAGUUUGAAAUAAUUGUCGAAU-- ------...((((((.-((......)).((((((....)))))).........))))))((((((...--...(((-.....)))...(((....)))....)))))).-.((((.....(((((----(.((..(.....)..)))))))).))))....-- ( -30.70, z-score = -2.04, R) >droAna3.scaffold_13340 9172340 145 - 23697760 -------AGUGCAGCA-GUAUUAAGCCAGACACUUUAUGAUAUCGGUAUAUCAGCUUUGGGCAGCAAAACAAACAG-CCACCCUGUAUACCCGUUC---ACGUUUAAAUAAUUUCCAAAUUUAAU----AUUACAAAAAUUCACUUCAAAGUGUAUCAUAU-- -------(((((.((.-.......))..).)))).(((((((..((((((.(((.....(((.............)-))...))))))))).....---..........................----............((((....))))))))))).-- ( -22.02, z-score = -0.21, R) >dp4.chr2 6488040 160 - 30794189 UGUGGGGGAUGCAGGAUACAGGAUGCCAGACAUUUUACGAUGUCGAUAAAUCCUGUGCAUUAUCGAUUUUGAAUCCUGUAUCGAUGCAAGCGGCCCAGAAAACUCCUAAUAGAAAAUUACUUACU--UAGCCCUUGCGAGCAA-GUAAAAAUCCACCUUGAAA .(((((.(((((((((((((((((....((((((....)))))).....)))))))......(((....)))))))))))))..((((((.(((..((.........................))--..))))))))).....-.......)))))....... ( -46.91, z-score = -2.99, R) >droPer1.super_3 299867 163 - 7375914 UGUGGGGGUUGCAGGAUGCAGGAUGCCAGACAUUUUACGAUGUCGAUAAAUCCUGUGCAUUAUCGAUUUUGAUUCCUGUAUCGAUGCAAGCGGCCCAUAAAACUCCUAAUAGAAAAUUACUUAAUGGCGGCUAUCGAGAAUAAUGUGAAAAAACCCCUAGAAA (.((((((((((((((.(((((((....((((((....)))))).....))))))).....((((....)))))))))).(((((...(((.(((..............................))).))))))))..............)))))))).).. ( -48.11, z-score = -2.60, R) >consensus ______AGUUGCAGCA_GGAUAAAGCCAGACACUUUAUGAUGUCGAUAAAUCAGCUGCAGGUAAUACCACGAUUCC_ACAAAGGGAAAAGUGGG____AGAAAUUAUAAUAGACAUACAUUUAUU____UUCAUUCAAUCAAAUUGAAAUAAUUGUCAAAU__ ........(((((((.............((((........))))((....)).)))))))....................................................................................................... ( -9.68 = -9.58 + -0.11)

| Location | 5,014,246 – 5,014,390 |
|---|---|
| Length | 144 |
| Sequences | 8 |
| Columns | 163 |
| Reading direction | reverse |
| Mean pairwise identity | 60.50 |
| Shannon entropy | 0.78227 |
| G+C content | 0.36093 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -9.19 |
| Energy contribution | -9.85 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.769874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
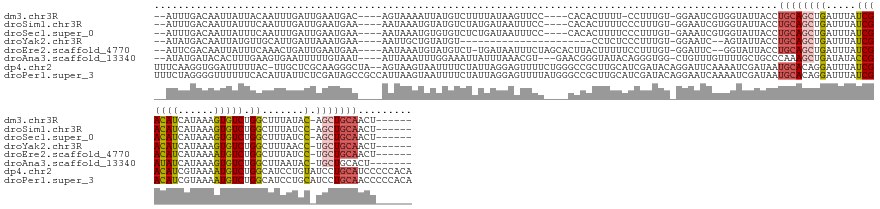
>dm3.chr3R 5014246 144 - 27905053 --AUUUGACAAUUAUUACAAUUUGAUUGAAUGAC----AGUAAAAUUAUGUCUUUUAUAAGUUCC----CACACUUUU-CCUUUGU-GGAAUCGUGGUAUUACCUGCAGCUGAUUUAUCGACAUCAUAAAGUGUCUGGCUUUAUAC-AGCUGCAACU------ --......((((((........))))))...(((----(.........))))............(----(((....((-((.....-))))..)))).......((((((((.......(....)(((((((.....))))))).)-)))))))...------ ( -31.80, z-score = -1.72, R) >droSim1.chr3R 11219686 145 + 27517382 --AUUUGACAAUUAUUUCAAUUUGAUUGAAUGAA----AAUAAAUGUAUGUCUAUGAUAAUUUCC----CACACUUUUCCCUUUGU-GGAAUCGUGGUAUUACCUGCAGCUGAUUUAUCGACAUCAUAAAGUGUCUGGCUUUAUCC-AGCUGCAACU------ --......((((((........))))))......----.......(((...(((((((......(----((((..........)))-)).)))))))...))).((((((((((...(((((((......))))).))....)).)-)))))))...------ ( -30.90, z-score = -1.52, R) >droSec1.super_0 4252801 145 + 21120651 --AUUUGACAAUUAUUUCAAUUUGAUUGAAUGAA----AAUAAAUGUGUGUCUCUGAUAAUUUCC----CACACUUUUCCCUUUGU-GAAAUCGUGGUAUUACCUGCAGCUGAUUUAUCGACAUCAUAAAGUGUCUGGCUUUAUCC-AGCUGCAACU------ --((((.((((((((((..((((....))))..)----)))))..(((((...............----)))))........))))-.))))...((.....))((((((((((...(((((((......))))).))....)).)-)))))))...------ ( -28.26, z-score = -0.97, R) >droYak2.chr3R 9063382 125 - 28832112 --AUAUGACAAUUAUGUUGCAUUGAUUAAAUGAA----AAUUGCUGUAUGU----------------------CCUCUCCCUUUGU-GGAAUC--AGUAUUACCUGCAGCUGAUUUAUCGACAUCAUAAAGUGUCUGGCUUUAACC-UGCUGCAACU------ --....((((....)))).((((.....))))..----...(((((....(----------------------((.(.......).-)))..)--)))).....((((((.........(((((......))))).((......))-.))))))...------ ( -24.80, z-score = -0.74, R) >droEre2.scaffold_4770 1159364 146 + 17746568 --AUUCGACAAUUAUUUCAAACUGAUUGAAUGAA----AAUAAAUGUAUGUCU-UGAUAAUUUCUAGCACUUACUUUUUCCUUUGU-GGAUUC--GGUAUUACCUGCAGCUGAUUUAUCGACAUCAUAAAAUGUCUGGCUUUAUCC-UGCUGCAACU------ --..((.((((....(((((.....))))).(((----((.....(((.((((-.((.....)).)).)).))).)))))..))))-.))...--((.....))((((((.(((...(((((((......))))).))....))).-.))))))...------ ( -26.70, z-score = -1.14, R) >droAna3.scaffold_13340 9172340 145 + 23697760 --AUAUGAUACACUUUGAAGUGAAUUUUUGUAAU----AUUAAAUUUGGAAAUUAUUUAAACGU---GAACGGGUAUACAGGGUGG-CUGUUUGUUUUGCUGCCCAAAGCUGAUAUACCGAUAUCAUAAAGUGUCUGGCUUAAUAC-UGCUGCACU------- --.((((((((((((((((...(((((..(....----........)..))))).)))))).))---)....(((((((((((..(-(..........))..))).....)).))))))..))))))).(((((..(((.......-.))))))))------- ( -36.30, z-score = -1.99, R) >dp4.chr2 6488040 160 + 30794189 UUUCAAGGUGGAUUUUUAC-UUGCUCGCAAGGGCUA--AGUAAGUAAUUUUCUAUUAGGAGUUUUCUGGGCCGCUUGCAUCGAUACAGGAUUCAAAAUCGAUAAUGCACAGGAUUUAUCGACAUCGUAAAAUGUCUGGCAUCCUGUAUCCUGCAUCCCCCACA .(((..((((((..(((((-((((((....)))).)--))))))......))))))..))).....((((.....((((..(((((((((((((.....(((((..........)))))(((((......)))))))).)))))))))).))))...)))).. ( -50.00, z-score = -3.19, R) >droPer1.super_3 299867 163 + 7375914 UUUCUAGGGGUUUUUUCACAUUAUUCUCGAUAGCCGCCAUUAAGUAAUUUUCUAUUAGGAGUUUUAUGGGCCGCUUGCAUCGAUACAGGAAUCAAAAUCGAUAAUGCACAGGAUUUAUCGACAUCGUAAAAUGUCUGGCAUCCUGCAUCCUGCAACCCCCACA ......((((((..............((((((((.(((.....((((..((((....))))..)))).))).)))...)))))..((((((((......)))......((((((.(...(((((......)))))..).))))))..))))).)))))).... ( -43.70, z-score = -2.24, R) >consensus __AUUUGACAAUUAUUUCAAUUUGAUUGAAUGAA____AAUAAAUGUAUGUCUAUUAUAAGUUCC____CACACUUUUCCCUUUGU_GGAAUCGUGGUAUUACCUGCAGCUGAUUUAUCGACAUCAUAAAGUGUCUGGCUUUAUCC_UGCUGCAACU______ ........................................................................................................((((((.((....))(((((......))))).............))))))......... ( -9.19 = -9.85 + 0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:16 2011