
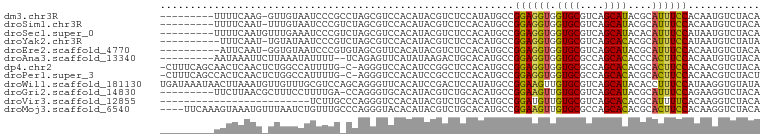
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 5,003,443 – 5,003,584 |
| Length | 141 |
| Max. P | 0.679857 |

| Location | 5,003,443 – 5,003,533 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.56 |
| Shannon entropy | 0.62239 |
| G+C content | 0.49314 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -11.46 |
| Energy contribution | -10.91 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5003443 90 + 27905053 ---------UUUUCAAG-GUUGUAAUCCCGCCUAGCGUCCACAUACGUCUCCAUAUGCCGGAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACA ---------......((-((.(.....).))))...((..((((....((((.......))))(((((((((......)))))...)))).))))..)). ( -20.80, z-score = -0.48, R) >droSim1.chr3R 11208485 90 - 27517382 ---------UUUUCAAU-UUUGUAAUCCCGUCUAGCGUCCACAUACGUCUCCACAUGCCGGAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACA ---------........-..................((..((((....((((.......))))(((((((((......)))))...)))).))))..)). ( -18.10, z-score = -0.56, R) >droSec1.super_0 4242061 91 - 21120651 ---------UUUUCAAUGUUUGAAAUCCCGUCUAGCGUCCACAUACGUCUCCACAUGCCGGAGGUGGUGCGUCAGCAUACACAUUUCCAUAAUGUCUACA ---------.(((((.....))))).........((((......))))....((((...(((((((((((....))))...)))))))...))))..... ( -16.70, z-score = 0.20, R) >droYak2.chr3R 9051482 89 + 28832112 ----------UUUCAAU-UGUAUAAUCCCGUCUAGCGUCCACAUACGUCUCCACAUGCCGGAGGUGGUGCGUCAGCACACGCAUUUCCAUAAUGUCUAUA ----------.......-................((((.......(..((((.......))))..)((((....)))))))).................. ( -18.80, z-score = -0.68, R) >droEre2.scaffold_4770 1148295 89 - 17746568 ----------AUUCAAU-GGUGUAAUCCCGUGUAGCGUUCACAUACGUCUCCACAUGCCGGAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACA ----------.....((-((.(....)))))((((((((.........((((.......))))(((((((((......)))))...)))))))).)))). ( -23.00, z-score = -0.41, R) >droAna3.scaffold_13340 9159750 89 - 23697760 ---------AAUAAAUUCUUAAAUAUUUU--UCAGAGUUCAUAUAAGACUGCACAUGCCGGAGGUGGUGCGCCAGCACACCCACUUCCACAAUGUGUACA ---------....((((((.(((....))--).))))))..........(((((((...(((((((((((....)))...))))))))...))))))).. ( -26.80, z-score = -3.60, R) >dp4.chr2 6474620 97 - 30794189 -CUUUCAGCAACUCAACUCUGGCCAUUUUG-C-AGGGUCCACAUCCGGCUCCACAUGCCGGAGGUGGUGCGCCAGCACACGCACUUCCACAACGUCUACA -......((......((((((.........-)-))))).(((.((((((.......)))))).)))((((....))))..)).................. ( -28.90, z-score = -1.26, R) >droPer1.super_3 286068 97 - 7375914 -CUUUCAGCCACUCAACUCUGGCCAUUUUG-C-AGGGUCCACAUCCGCCUCCACAUGCCGGAGGUGGUGCGCCAGCACACGCACUUCCACAACGUCUACU -......((((........)))).......-.-.(((........(((((((.......)))))))(((((........))))).)))............ ( -25.80, z-score = -0.58, R) >droWil1.scaffold_181130 8371959 100 - 16660200 UGAUAAAUAACUUAAAUGUUGUUUGCGUCCAGCAGGGUUCACAUCCGACUCCAUAUGCCGGAAGUUGUGCGUCAGCAUACACCUUUCCAUAAGGUGUAUA .(.((((((((......))))))))).(((.(((.((.((......))..))...))).))).((((.....))))(((((((((.....))))))))). ( -29.00, z-score = -2.44, R) >droGri2.scaffold_14830 2393698 90 + 6267026 ---------UUCUUAACGCUUUCCUUUUGA-CCAGGGUGCACAUACGUCUGCACAUGCCGGAAGUUGUGCGUCAGCAUACGCAUUUCCAGAAGGUCUACA ---------...................((-((.(((((((........)))))...))(((((((((((....)))))...))))))....)))).... ( -23.70, z-score = -0.84, R) >droVir3.scaffold_12855 8709299 75 + 10161210 -------------------------UCUUGCCCAGGGUCCACAUACGUCUGCACAUGCCGGAUGUUGUGCGUCAGCACACGCAUUUUCACAAGGUCUACA -------------------------(((((.....((..((((.((((((((....).)))))))))))..)).((....)).......)))))...... ( -17.10, z-score = 0.64, R) >droMoj3.scaffold_6540 23893762 96 + 34148556 ----UUCAAAGUAAAUGUUUAAUCUGUUUGCCCAGGGUACACAUACGUCUGCACAUGCCGGAAGUUGUGCGCCAGCACACGCACUUCCACAAGGUCUACA ----......(((..(((....((((......))))..)))..)))((..((.(.((..(((((((((((....)))))...)))))).)).)))..)). ( -22.30, z-score = 0.21, R) >consensus _________AUUUCAAU_UUUGUAAUCUCG_CCAGGGUCCACAUACGUCUCCACAUGCCGGAGGUGGUGCGUCAGCACACGCAUUUCCACAAUGUCUACA ...........................................................((((((.((((....))))....))))))............ (-11.46 = -10.91 + -0.55)

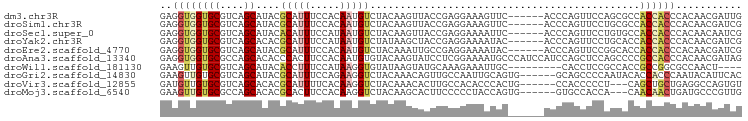
| Location | 5,003,493 – 5,003,584 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 67.05 |
| Shannon entropy | 0.68214 |
| G+C content | 0.52911 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -7.27 |
| Energy contribution | -7.73 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 5003493 91 + 27905053 GAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACAAGUUACCGAGGAAAGUUC------ACCCAGUUCCAGCGCCACCACCCACAACGAUUG ..((((((((((..((((.............))))..))..........((((.....------......)))).)))))))).............. ( -21.22, z-score = -1.24, R) >droSim1.chr3R 11208535 91 - 27517382 GAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACAAGUUACCGAGGAAAGUUC------ACCCAGUUCCUGCGCCACCACCCACAACGAUCG ..((((((((((..((((.............))))..)).........(((((.....------......))))))))))))).............. ( -21.82, z-score = -1.15, R) >droSec1.super_0 4242112 91 - 21120651 GAGGUGGUGCGUCAGCAUACACAUUUCCAUAAUGUCUACAAGUUACCGAGGAAAAUUC------ACCCAGUUCCUGUGCCACCACCCACAACAAUCG ..(((((..((..(((.((.(((((.....))))).))...)))..).(((((.....------......))))))..))))).............. ( -19.70, z-score = -1.37, R) >droYak2.chr3R 9051531 91 + 28832112 GAGGUGGUGCGUCAGCACACGCAUUUCCAUAAUGUCUAUAAGCUACCGAGGAAAAUAC------ACCCAGUUCCUGCACCACCACCCACAACGAUCG ..(((((((((..(((....(((((.....)))))......)))..).(((((.....------......))))))))))))).............. ( -24.80, z-score = -3.03, R) >droEre2.scaffold_4770 1148344 91 - 17746568 GAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACAAAUUGCCGAGGAAAAUAC------ACCCAGUUCCGGCACCACCACCCACAACGAUCG ..((((((((....((....)).(((((.((((........))))....)))))....------...........)))))))).............. ( -23.50, z-score = -1.87, R) >droAna3.scaffold_13340 9159799 97 - 23697760 GAGGUGGUGCGCCAGCACACCCACUUCCACAAUGUGUACAAGUAUCCUCGGAAAAUGCCCAUCCAUCCAGCUCCAGCCCCGCCACCCACAACGAUAG ..(((((.((...(((......((((..((.....))..))))......(((.........))).....)))...)).))))).............. ( -19.40, z-score = -0.46, R) >droWil1.scaffold_181130 8372019 84 - 16660200 GAAGUUGUGCGUCAGCAUACACCUUUCCAUAAGGUGUAUAAGUAUGCAAAGAAAUUGC---------CACCUCCGCCACCGCCGGCGCCAACU---- ..(((((.(((((.(((((((((((.....)))))))))......((((.....))))---------.............)).))))))))))---- ( -29.60, z-score = -4.56, R) >droGri2.scaffold_14830 2393748 91 + 6267026 GAAGUUGUGCGUCAGCAUACGCAUUUCCAGAAGGUCUACAAACAGUUGCCAAUUGCAGUG------GCAGCCCCAAUACACCACCCAAUACAUUCAC ((((..((((((......))))))..).....(((.((......(((((((.......))------))))).....)).)))..........))).. ( -19.40, z-score = -1.14, R) >droVir3.scaffold_12855 8709334 88 + 10161210 GAUGUUGUGCGUCAGCACACGCAUUUUCACAAGGUCUACAAACACUUGCCACACCCACUG------CCACCCCCU---CAGCUGCUGAGGCCAGUGU ((((((((((....))))).)))))....((((((......)).)))).......(((((------......(((---(((...)))))).))))). ( -23.30, z-score = -0.46, R) >droMoj3.scaffold_6540 23893818 88 + 34148556 GAAGUUGUGCGCCAGCACACGCACUUCCACAAGGUCUACAAGCACUUCCCCCUACCAGUG------GUGCCACCA---CAACAACUGAUGCCCGUUG ((((((((((....))))).((....((....)).......))))))).........(((------(.....)))---)..((((........)))) ( -18.80, z-score = 0.37, R) >consensus GAGGUGGUGCGUCAGCAUACGCAUUUCCACAAUGUCUACAAGUAACCGAGGAAAAUAC______ACCCAGCUCCAGCACCACCACCCACAACGAUCG ..((((((((....))....(((((.....))))).............................................))))))........... ( -7.27 = -7.73 + 0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:12 2011