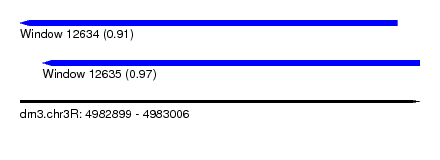
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,982,899 – 4,983,006 |
| Length | 107 |
| Max. P | 0.973585 |

| Location | 4,982,899 – 4,983,000 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Shannon entropy | 0.44734 |
| G+C content | 0.58074 |
| Mean single sequence MFE | -40.01 |
| Consensus MFE | -27.34 |
| Energy contribution | -26.02 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
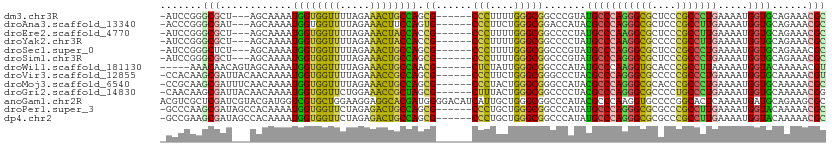
>dm3.chr3R 4982899 101 - 27905053 -AUCCGGGCGCU---AGCAAAAUGGUGGUUUUAGAAACUGCCAGCG------CCCUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAGAAACGC -...(((((...---.......(((..(((.....)))..))).((------(((....))))))))))...((((((((((....))))))).....))).......... ( -42.80, z-score = -1.26, R) >droAna3.scaffold_13340 9140348 101 + 23697760 -ACCCGGGCGAU---AGCAAAAUGGUGGUUUUAGAAACUUCCAGUG------CCCUUCUGGGCGGACCAUACGCCCAGGGCGCUCCCGCCUUGAAAAUGGUGCAGAAACGC -..(.(((((..---(((....(((.((((.....)))).)))..(------((((...(((((.......)))))))))))))..))))).).................. ( -36.70, z-score = -0.96, R) >droEre2.scaffold_4770 1127369 101 + 17746568 -AUCCGGGCGCU---AGCAAAAUGGUGGUUUUAGAAACUACCACCG------CCCUUUUGGGCGGCCCCUAUGCCCAAGGCGCUCCCGCCUUGAAAAUGGUGCAGAAACGC -......(((((---((((...((((((((.....))))))))(((------(((....))))))......))).(((((((....)))))))....))))))........ ( -43.10, z-score = -2.83, R) >droYak2.chr3R 9030397 101 - 28832112 -AUCCGGGCGCU---AGCAAAAUGGUGGUUUUAGAAACUACCACCG------CCCUUUUGGGCGGCCCCUAUGCCCAAGGCGCCCCCGCCUUGAAAAUGGUGCAGAAACGC -......(((((---((((...((((((((.....))))))))(((------(((....))))))......))).(((((((....)))))))....))))))........ ( -43.10, z-score = -2.68, R) >droSec1.super_0 4221542 101 + 21120651 -AUCCGGGCUCU---AGCAAAAUGGUGGUUUUAGAAACUGCCAGCG------CCCUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAGAAACGC -...(((((...---.......(((..(((.....)))..))).((------(((....))))))))))...((((((((((....))))))).....))).......... ( -42.30, z-score = -1.52, R) >droSim1.chr3R 11186609 101 + 27517382 -AUCCGGGCGCU---AGCAAAAUGGUGGUUUUAGAAACUGCCAGCG------CCCUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAGAAACGC -...(((((...---.......(((..(((.....)))..))).((------(((....))))))))))...((((((((((....))))))).....))).......... ( -42.80, z-score = -1.26, R) >droWil1.scaffold_181130 8349789 100 + 16660200 -----AAACAACAGUAGCAAAAUGGUGGUUUUAGAAACUGCCAACG------CUCUAUUGGGCGGCCCAUAUGCCCAAGGUGCACCCGCCUUAAAAAUGGUACAAAAACGU -----........(((.((...(((..(((.....)))..)))..(------(.((.(((((((.......))))))))).))..............)).)))........ ( -26.70, z-score = -0.39, R) >droVir3.scaffold_12855 8682725 104 - 10161210 -CCACAAGCGAUUACAACAAAAUGGUGGUUUUAGAAACCGCCAGCG------CCCUUCUGGGCGGGCCCUACGCCCAGGGCGCCCCCGCCCUGAAAAUGGUGCAAAAACGU -......(((............((((((((.....))))))))(((------((.(((.(((((((((((......)))))....)))))).)))...))))).....))) ( -43.80, z-score = -2.94, R) >droMoj3.scaffold_6540 23869889 104 - 34148556 -CCGCAAGCGAUUUCAACAAAAUGGUGGUUUUAGAAACUGCCAGCG------CCCUACUGGGCGGGCCAUACGCCCAGGGCGCACCCGCCCUGAAAAUGGUGCAAAAACGC -..(((..((.(((((......(((..(((.....)))..)))(((------((((...(((((.......))))))))))))........))))).)).)))........ ( -40.90, z-score = -1.96, R) >droGri2.scaffold_14830 2371305 104 - 6267026 -CAACAAGCGAUUACAACAAAAUGGUGGUUCUGGAAACCGCUAGCG------CUUUACUGGGCGGCCCCUACGCCCAGGGCGCCCCUGCCCUGAAAAUGGUGCAAAAACGG -.......((............((((((((.....))))))))(((------((...(((((((.......))))))))))))...(((((.......)).)))....)). ( -35.60, z-score = -0.89, R) >anoGam1.chr2R 47960496 111 - 62725911 ACGUCGCUCGAUCGUACGAUGGUCGUGCUGGAAGGAGGCACGAUGGGGACAUCAUUGCUGGGCGGCCCAUACGCCCAAGGUGCCCCGGCACUCAAAAUGAUGCAGAAGCGC ....(((((((((((...))))))(((((((.....(((((((((....)))).....((((((.......))))))..)))))))))))).............).)))). ( -47.10, z-score = -1.82, R) >droPer1.super_3 253400 104 + 7375914 -GCCCAAGCGAUAGCCACAAAAUGGUGGUUCUAGAGACUGCCAGCG------CCCUGCUGGGCGGCCCAUAUGCCCAGGGCGCGCCCGCCUUGAAAAUGGUACAAAAACGC -......(((...((.......(((..(((.....)))..)))(((------((((...(((((.......))))))))))))))..(((........))).......))) ( -37.60, z-score = -0.47, R) >dp4.chr2 6446231 104 + 30794189 -GCCGAAGCGAUAGCCACAAAAUGGUGGUUCUAGAGACUGCCAGCG------CCCUGCUGGGCGGCCCAUAUGCCCAGGGCGCGCCCGCCUUGAAAAUGGUACAAAAACGC -......(((...((.......(((..(((.....)))..)))(((------((((...(((((.......))))))))))))))..(((........))).......))) ( -37.60, z-score = -0.53, R) >consensus _ACCCGAGCGAU___AGCAAAAUGGUGGUUUUAGAAACUGCCAGCG______CCCUUCUGGGCGGCCCAUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAGAAACGC .......(((............((((((((.....))))))))..........((....(((((.......)))))((((((....))))))......))........))) (-27.34 = -26.02 + -1.32)

| Location | 4,982,905 – 4,983,006 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Shannon entropy | 0.49579 |
| G+C content | 0.58676 |
| Mean single sequence MFE | -41.89 |
| Consensus MFE | -25.92 |
| Energy contribution | -24.90 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4982905 101 - 27905053 ----UCGCAGAUCCGGGCGCUAGCAAAAUGGUGGUUUUAGAAACUGCCA--------GCGCC-CUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAG ----..(((....(((((..........(((..(((.....)))..)))--------.((((-(....))))))))))......(((((((....))))))).......))).. ( -45.40, z-score = -1.89, R) >droAna3.scaffold_13340 9140354 101 + 23697760 ----UUGCAGACCCGGGCGAUAGCAAAAUGGUGGUUUUAGAAACUUCCA--------GUGCC-CUUCUGGGCGGACCAUACGCCCAGGGCGCUCCCGCCUUGAAAAUGGUGCAG ----.((((...(((((((..(((....(((.((((.....)))).)))--------..(((-((...(((((.......)))))))))))))..))))).......)))))). ( -39.01, z-score = -1.54, R) >droEre2.scaffold_4770 1127375 101 + 17746568 ----UCGCUGAUCCGGGCGCUAGCAAAAUGGUGGUUUUAGAAACUACCA--------CCGCC-CUUUUGGGCGGCCCCUAUGCCCAAGGCGCUCCCGCCUUGAAAAUGGUGCAG ----..((....(((((((.(((.....((((((((.....))))))))--------(((((-(....))))))...))))))))((((((....))))))......)).)).. ( -43.90, z-score = -3.01, R) >droYak2.chr3R 9030403 101 - 28832112 ----UCGCUGAUCCGGGCGCUAGCAAAAUGGUGGUUUUAGAAACUACCA--------CCGCC-CUUUUGGGCGGCCCCUAUGCCCAAGGCGCCCCCGCCUUGAAAAUGGUGCAG ----..((....(((((((.(((.....((((((((.....))))))))--------(((((-(....))))))...))))))))((((((....))))))......)).)).. ( -43.90, z-score = -2.90, R) >droSec1.super_0 4221548 101 + 21120651 ----UCGCAGAUCCGGGCUCUAGCAAAAUGGUGGUUUUAGAAACUGCCA--------GCGCC-CUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAG ----..(((....(((((..........(((..(((.....)))..)))--------.((((-(....))))))))))......(((((((....))))))).......))).. ( -44.90, z-score = -2.16, R) >droSim1.chr3R 11186615 101 + 27517382 ----UCGCAGAUCCGGGCGCUAGCAAAAUGGUGGUUUUAGAAACUGCCA--------GCGCC-CUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAG ----..(((....(((((..........(((..(((.....)))..)))--------.((((-(....))))))))))......(((((((....))))))).......))).. ( -45.40, z-score = -1.89, R) >droWil1.scaffold_181130 8349795 100 + 16660200 ----UAACAGAAAC-AACAGUAGCAAAAUGGUGGUUUUAGAAACUGCCA--------ACGCU-CUAUUGGGCGGCCCAUAUGCCCAAGGUGCACCCGCCUUAAAAAUGGUACAA ----..........-....(((.((...(((..(((.....)))..)))--------..((.-((.(((((((.......))))))))).))..............)).))).. ( -26.70, z-score = -0.45, R) >droVir3.scaffold_12855 8682731 104 - 10161210 -UCGGCACCACAAGCGAUUACAACAAAAUGGUGGUUUUAGAAACCGCCA--------GCGCC-CUUCUGGGCGGGCCCUACGCCCAGGGCGCCCCCGCCCUGAAAAUGGUGCAA -...((((((...(((............((((((((.....))))))))--------.((((-(....))))).......))).(((((((....)))))))....)))))).. ( -48.70, z-score = -3.54, R) >droMoj3.scaffold_6540 23869895 104 - 34148556 -UCGGGACCGCAAGCGAUUUCAACAAAAUGGUGGUUUUAGAAACUGCCA--------GCGCC-CUACUGGGCGGGCCAUACGCCCAGGGCGCACCCGCCCUGAAAAUGGUGCAA -..(((.((....).)............(((..(((.....)))..)))--------(((((-((...(((((.......)))))))))))).)))((((.......)).)).. ( -43.40, z-score = -1.90, R) >droGri2.scaffold_14830 2371311 104 - 6267026 -UCGGCACAACAAGCGAUUACAACAAAAUGGUGGUUCUGGAAACCGCUA--------GCGCU-UUACUGGGCGGCCCCUACGCCCAGGGCGCCCCUGCCCUGAAAAUGGUGCAA -...((((...(((((............((((((((.....))))))))--------.))))-)....(((((.......)))))((((((....)))))).......)))).. ( -37.72, z-score = -0.84, R) >anoGam1.chr2R 47960502 111 - 62725911 ---UCGGCAACGUCGCUCGAUCGUACGAUGGUCGUGCUGGAAGGAGGCACGAUGGGGACAUCAUUGCUGGGCGGCCCAUACGCCCAAGGUGCCCCGGCACUCAAAAUGAUGCAG ---.((....))..((((((((((...))))))(((((((.....(((((((((....)))).....((((((.......))))))..)))))))))))).......)).)).. ( -48.40, z-score = -2.23, R) >droPer1.super_3 253406 104 + 7375914 UCACAAGCCCAAGC-GAUAGCCACAAAAUGGUGGUUCUAGAGACUGCCA--------GCGCC-CUGCUGGGCGGCCCAUAUGCCCAGGGCGCGCCCGCCUUGAAAAUGGUACAA ......((((((((-(...((.......(((..(((.....)))..)))--------(((((-((...(((((.......)))))))))))))).)).)))).....))).... ( -37.60, z-score = -0.44, R) >dp4.chr2 6446237 104 + 30794189 UCACAGGCCGAAGC-GAUAGCCACAAAAUGGUGGUUCUAGAGACUGCCA--------GCGCC-CUGCUGGGCGGCCCAUAUGCCCAGGGCGCGCCCGCCUUGAAAAUGGUACAA (((.((((....((-.............(((..(((.....)))..)))--------(((((-((...(((((.......))))))))))))))..)))))))........... ( -39.50, z-score = -0.63, R) >consensus ____UCGCAGAACCGGACGCUAGCAAAAUGGUGGUUUUAGAAACUGCCA________GCGCC_CUUCUGGGCGGCCCAUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAG ............................((((((((.....))))))))..........(((......(((((.......)))))((((((....))))))......))).... (-25.92 = -24.90 + -1.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:08 2011