
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,971,777 – 4,971,886 |
| Length | 109 |
| Max. P | 0.852481 |

| Location | 4,971,777 – 4,971,886 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 67.51 |
| Shannon entropy | 0.57631 |
| G+C content | 0.49860 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -9.51 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
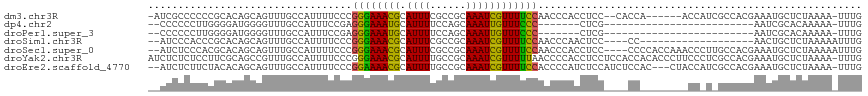
>dm3.chr3R 4971777 109 + 27905053 -AUCGCCCCCCGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCACCUCC--CACCA------ACCAUCGCCACGAAAUGCUCUAAAA-UUUG -...((.....))......((((.((........)).))))((((((((..((.......................--.....------......))..)))))))).......-.... ( -17.28, z-score = -0.96, R) >dp4.chr2 6435450 84 - 30794189 --CCCCCCUUGGGGAUGGGGUUUGCCAUUUCCGAGGGAAAUGCAUUUUCCAGCAAAUUGUUUCCC-------CUCG-------------------------AAUCGCACAAAAA-UUUG --...(((((((..((((......))))..)))))))...(((........)))...........-------....-------------------------.............-.... ( -24.10, z-score = -0.38, R) >droPer1.super_3 242568 84 - 7375914 --CCCCCCUUGGGGAUGGGGUUUGCCAUUUCCGAGGGAAAUGCAUUUUCCAGCAAAUUGUUUCCC-------CUCG-------------------------AAUCGCACAAAAA-UUUG --...(((((((..((((......))))..)))))))...(((........)))...........-------....-------------------------.............-.... ( -24.10, z-score = -0.38, R) >droSim1.chr3R 11174983 94 - 27517382 --AUCCCACCCGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCAACUCC----CC-------------------AACUGCUCUAAAAAUUUG --.............(((((((............((((((((.((((......))))))))))))...........----..-------------------)))))))........... ( -16.71, z-score = -2.22, R) >droSec1.super_0 4210643 113 - 21120651 --AUCUCCCACGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCACCUCC----CCCCACCAAACCCUUGCCACGAAAUGCUCUAAAAAUUUG --.................((((.((........)).))))((((((((..((((...((((..............----.......))))..))))..))))))))............ ( -19.70, z-score = -2.53, R) >droYak2.chr3R 9019106 118 + 28832112 AUCUCUCUCCUUCGCAGCCGUUUGCCAUUUUCCCGGGAAACGCAUUUUGCCGCAAAUCGUUUUUAACCCCACCUCCUCCACCACACCCUUCCCUCGCCACGAAAUGCUCUAAAA-UUUG ...................((((.((........)).))))((((((((..((..........................................))..)))))))).......-.... ( -14.35, z-score = -0.71, R) >droEre2.scaffold_4770 1116239 113 - 17746568 --AUCUCUUCUACACAGCAGUUUGCCAUUUUCCCGGAAAACGCAUUUUGCCGCAAAUCGUUUUCCACCCCAUCUCCAUCUCCAC---CUACCAUCGCCACGAAAUGCUCUAAAA-UUUG --.............((((...............((((((((.((((......))))))))))))...................---......(((...)))..))))......-.... ( -14.40, z-score = -1.85, R) >consensus __AUCCCCCCCGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUUGCCGCAAAUCGUUUUCCAACCCACCUCC____CC________CC_U_GCCACGAAAUGCUCUAAAA_UUUG ..................................((((((((.((((......))))))))))))...................................................... ( -9.51 = -8.80 + -0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:00:02 2011