
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,935,733 – 4,935,823 |
| Length | 90 |
| Max. P | 0.545182 |

| Location | 4,935,733 – 4,935,823 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Shannon entropy | 0.42135 |
| G+C content | 0.36526 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
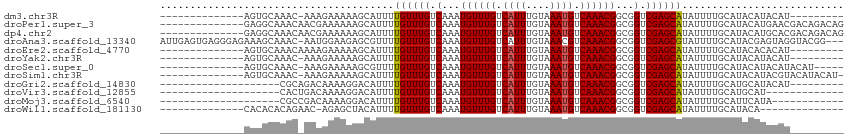
>dm3.chr3R 4935733 90 - 27905053 --------------AGUGCAAAC-AAAGAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUACAU--------- --------------.(((((((.-...(......)....((((((.(...((((((.((((....)))).))))))...).))))))...)))))))........--------- ( -20.10, z-score = -1.15, R) >droPer1.super_3 200060 100 + 7375914 --------------GAGGCAAACAACGAAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUGAACGACAGACAG --------------..(((((((((.............)))))))))...((((((((.((..(..(((.((((..((......))....)))))))..)..)).)))))))). ( -25.72, z-score = -1.84, R) >dp4.chr2 6393109 100 + 30794189 --------------GAGGCAAACAACGAAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUGCACGACAGACAG --------------..(((((((((.............)))))))))...((((((((...((((..((((....)))).....(((.....))).)))).....)))))))). ( -25.02, z-score = -1.33, R) >droAna3.scaffold_13340 9093657 110 + 23697760 AUUGAGUGAGGGAGAAAGCAAAC-AAUGGAAGAGCGUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAACGUCAAACGGCGGUCGAGCGUAUUUUGCAUACGAGUAGGUACGG--- ...((((((.(((.(..((((((-((............))))))))....).))).)))))).....((((....))))......(((((((.((......))))))))).--- ( -22.20, z-score = 0.75, R) >droEre2.scaffold_4770 1080287 91 + 17746568 --------------AGUGCAAACAAAAGAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACACACAU--------- --------------.(((((((.....(......)....((((((.(...((((((.((((....)))).))))))...).))))))...)))))))........--------- ( -20.10, z-score = -0.90, R) >droYak2.chr3R 8982762 90 - 28832112 --------------AGUGCAAAC-AAAGAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUACAU--------- --------------.(((((((.-...(......)....((((((.(...((((((.((((....)))).))))))...).))))))...)))))))........--------- ( -20.10, z-score = -1.15, R) >droSec1.super_0 4175176 94 + 21120651 --------------AGUGCAAAC-AAAGAAAAAGCGUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUACAUACAU----- --------------.(((((((.-...(......)....((((((.(...((((((.((((....)))).))))))...).))))))...)))))))............----- ( -20.10, z-score = -0.65, R) >droSim1.chr3R 11137844 98 + 27517382 --------------AGUGCAAAC-AAAGAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUACGUACAUACAU- --------------.(((((((.-...(......)....((((((.(...((((((.((((....)))).))))))...).))))))...)))))))................- ( -20.10, z-score = -0.40, R) >droGri2.scaffold_14830 2309824 85 - 6267026 --------------------CGCAGACAAAAGGACAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUGCAUACAU--------- --------------------.(((..(((((........((((((.(...((((((.((((....)))).))))))...).))))))..)))))..)))......--------- ( -21.70, z-score = -1.41, R) >droVir3.scaffold_12855 8617343 81 - 10161210 --------------------CACUGACAAAAGGACAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUGCAU------------- --------------------...((((.....((((.......))))...((((((.((((....)))).))))))...)))).(((.....)))......------------- ( -17.60, z-score = -0.24, R) >droMoj3.scaffold_6540 23810717 82 - 34148556 --------------------CGCCGACAAAAGGACAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUUCAUA------------ --------------------((((((((((..((...))..))))))....(((((.((((....)))).))))))))).....(((.....))).......------------ ( -23.10, z-score = -2.48, R) >droWil1.scaffold_181130 8294660 85 + 16660200 --------------CACACACAGAAC-AGAGCUACAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACA-------------- --------------............-...((.......((((((.(...((((((.((((....)))).))))))...).))))))......)).....-------------- ( -17.82, z-score = -0.45, R) >consensus ______________AGUGCAAAC_AAAAAAAAAGCAUUUUGUUUGUCAAAUGUUUGUCAUUUGUAAAUGUCAAACGGCGGUCGAGCAUAUUUUGCAUACAUACAU_________ .......................................((((((.(...((((((.((((....)))).))))))...).))))))........................... (-15.07 = -15.08 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:58 2011