
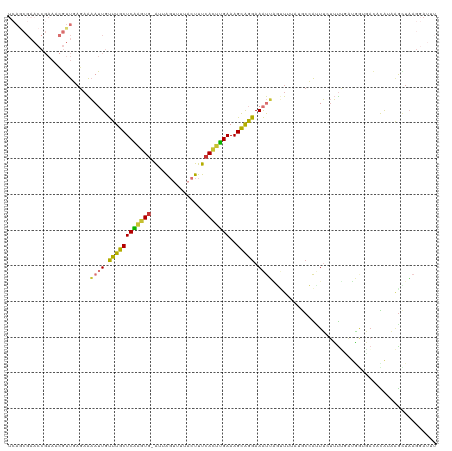
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,919,025 – 4,919,183 |
| Length | 158 |
| Max. P | 0.605959 |

| Location | 4,919,025 – 4,919,144 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.89 |
| Shannon entropy | 0.71454 |
| G+C content | 0.38394 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -10.21 |
| Energy contribution | -9.59 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4919025 119 - 27905053 UCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUUGUUUUAGGCCAUUUCUUUUCAUGGUGACUCAUAAGCUAAGGAUUU ((((((........))))))..(((((.((((((((((((-(..((.....)).))))))))..))))).))))).(((((((((((........)))))..((....)))))))).... ( -26.10, z-score = -0.47, R) >droSim1.chr3R 11121229 119 + 27517382 UCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUUAUGCCAUUUCUUUUCAUGGUGACUCAUAAGCUAAGGAUUU ((((((........))))))........((((((((((((-(..((.....)).))))))))..)))))...(((((((...(((((........))))).......)))))))...... ( -26.70, z-score = -0.98, R) >droSec1.super_0 4158624 119 + 21120651 UCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUUAUGCCAUUUCUUUUCAUGGUGACUCAUAAGCUAAGGAUUU ((((((........))))))........((((((((((((-(..((.....)).))))))))..)))))...(((((((...(((((........))))).......)))))))...... ( -26.70, z-score = -0.98, R) >droYak2.chr3R 8965820 119 - 28832112 UCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUACUUUUACUUUCACUUGAAAGCAGGAAAUUAGAUUUGUACCAUUUCUUUCGAUGGUGACGUAUACGCUAAGGAUUU ((((((........))))))........((((((((((((-(............))))))))..)))))...((((...(((((((((......)))))).)))......))))...... ( -26.10, z-score = -0.96, R) >droEre2.scaffold_4770 1062674 119 + 17746568 UCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUUAUGCCACUUAUUUUAAUGGUGACGUAUAAGUUAAGGAUUU ((((((........))))))...((((.((((((((((((-(..((.....)).))))))))..))))).))))............((((.(((((((....))).)))).))))..... ( -27.60, z-score = -1.83, R) >droAna3.scaffold_13340 9077235 111 + 23697760 -AAACGAACCGAAUCGUU----AAAUUGUUUACUCGGCUAUGGCAUUAUAACUUUCAUUUGAAAGUGGGAAAUGCAUUAUCUGGAAUUUAUUUGGGGAAACCAAUAAAAACUAACA---- -.(((((......)))))----.....((((..((((.....(((((...((((((....))))))....))))).....))))..........((....)).....)))).....---- ( -22.40, z-score = -1.72, R) >dp4.chr2 6374576 95 + 30794189 --------------UUUUAAGAGCAGUGCCUGCUCGAGUGUGGCGGC-AAACUUUCAUUCGAAAGCAGGAAGCGGAGU----GCUGCUGAUGCCGACCAAUAAACGCAGGAUUU------ --------------......((((((...))))))(.((.(((((((-(..(((((....))))).....(((((...----.)))))..))))).)))....)).).......------ ( -30.60, z-score = -0.88, R) >droPer1.super_3 182319 92 + 7375914 --------------UUUUAAGAGCAGUGCCUGCUCGUGUGUGGCGGC--AACUUUCAUUCGAAAGCAGGAAGCGGACU----GCUGAUG--GUCGACCAAUAAACGCUGGUUCU------ --------------......((((((...))))))((((.(((((((--....(((....)))(((((........))----)))....--)))).)))....)))).......------ ( -28.60, z-score = -0.47, R) >consensus UCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG_AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUUAUGCCAUUUCUUUUGAUGGUGACACAUAAGCUAAGGAUUU .......................((((.((((((((((((....((.....))..)))))))..))))).)))).............................................. (-10.21 = -9.59 + -0.62)


| Location | 4,919,065 – 4,919,183 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Shannon entropy | 0.34007 |
| G+C content | 0.32449 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -13.77 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
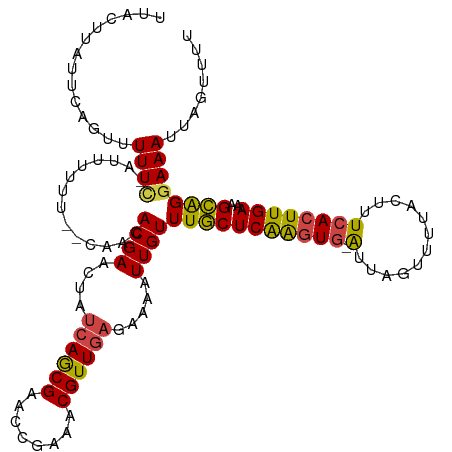
>dm3.chr3R 4919065 118 - 27905053 UUACUUAUUCAACUUUUC-AUUUUUUUGCAACACGAACUAUCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUUGUUUU .........(((.(((((-........((((.((((....((((((........)))))).....))))))))(((((((-(..((.....)).)))))))).....))))).))).... ( -26.10, z-score = -2.13, R) >droSim1.chr3R 11121269 119 + 27517382 UUACUUAUUCAGUUUUUCAAUUUUUUUGCAACACGAACUAUCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUU .....................((((((((....(((((..((((((........)))))).......))))).(((((((-(..((.....)).))))))))..))))))))........ ( -26.00, z-score = -1.82, R) >droSec1.super_0 4158664 118 + 21120651 UUACUUAUUCAGUUUUUC-AUUUUUUUGCAACACGAACUAUCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUU ..................-..((((((((....(((((..((((((........)))))).......))))).(((((((-(..((.....)).))))))))..))))))))........ ( -26.00, z-score = -1.90, R) >droYak2.chr3R 8965860 111 - 28832112 UUACUUUCUCAG-UUUUG-GUU------CAACACGAACUAUCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUACUUUUACUUUCACUUGAAAGCAGGAAAUUAGAUUU .....(((((((-(((((-(((------(................))))))))))..))))))((((.((((((((((((-(............))))))))..))))).))))...... ( -30.69, z-score = -3.48, R) >droEre2.scaffold_4770 1062714 112 + 17746568 UUACUUCUUCAGUUUUUU-UUU------CAACACGAACUAUCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG-AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUU .........((((((((.-.((------(.....)))...((((((........))))))))))))))((((((((((((-(..((.....)).))))))))..)))))........... ( -23.30, z-score = -1.14, R) >droAna3.scaffold_13340 9077271 113 + 23697760 UUACUUUUGAGUUAUUUCUAUCACGC--CAACACGAAACG-AAACGAACCGAAUCGUU----AAAUUGUUUACUCGGCUAUGGCAUUAUAACUUUCAUUUGAAAGUGGGAAAUGCAUUAU ..........((.((((((.....((--((...(((((((-((((((......)))))----...)))))...)))....))))......((((((....)))))).))))))))..... ( -23.70, z-score = -1.83, R) >consensus UUACUUAUUCAGUUUUUC_AUUUUUU__CAACACGAACUAUCAGCGAACCGAAACGUUGAGAAAAUUGUUUGCUCAAGUG_AUUAGUUUUACUUUCACUUGAAAGCAGGAAAUUAGUUUU ........................................((((((........))))))...((((.((((((((((((....((.....))..)))))))..))))).))))...... (-13.77 = -13.98 + 0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:56 2011