| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,909,104 – 4,909,210 |
| Length | 106 |
| Max. P | 0.864738 |

| Location | 4,909,104 – 4,909,210 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Shannon entropy | 0.41196 |
| G+C content | 0.47567 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
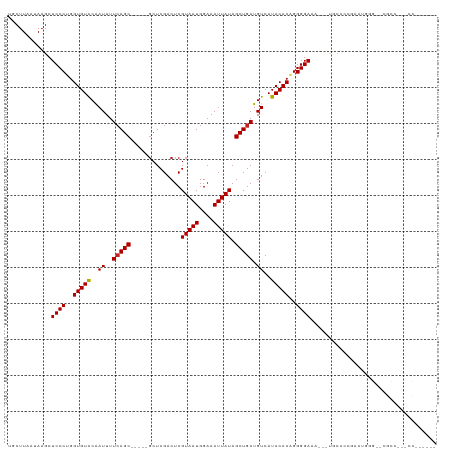
>dm3.chr3R 4909104 106 + 27905053 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAA---UGCUCCGACUGGGGUCGCCUUACCG----- .(((.....)))(((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))..---.(((((....)))))..........----- ( -27.80, z-score = -0.45, R) >droSim1.chr3R 11111430 106 - 27517382 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAA---UGCUCCGACUGGGGUCGCCUUACCG----- .(((.....)))(((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))..---.(((((....)))))..........----- ( -27.80, z-score = -0.45, R) >droSec1.super_0 4148870 85 - 21120651 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAA---UGCU-------------------------- .(((.....)))(((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))..---....-------------------------- ( -23.10, z-score = -1.76, R) >droYak2.chr3R 8955645 105 + 28832112 UGCUUAAUAAGCUCCCUGGUGUGCAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAGC---UGCCCCGAGUGGGA-CGCCCCACCA----- .........((((((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))))---).....(.(((((.-...)))))).----- ( -37.40, z-score = -2.16, R) >droEre2.scaffold_4770 1053008 106 - 17746568 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAA---UGCCCCGAGUGGGUUCGCCCUGCCA----- .((.......((((....(((((..((..(((((-----.........(((((....))))).))))).))..))))).((((...---..))))))))(((....))).))..----- ( -29.70, z-score = -0.37, R) >droAna3.scaffold_13340 9067244 111 - 23697760 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAA---UGCCCUGACUCGGGUCGCUUUUCCAUUACA ........(((((((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))..---.((((......)))).)))).......... ( -29.70, z-score = -1.20, R) >dp4.chr2 6361989 87 - 30794189 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGACU---CUGCCC------------------------ .(((.....)))(((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))..---......------------------------ ( -22.80, z-score = -1.15, R) >droPer1.super_3 169654 87 - 7375914 UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC-----GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGACU---CUGCCC------------------------ .(((.....)))(((((.(((((..((..(((((-----.........(((((....))))).))))).))..))))).)))))..---......------------------------ ( -22.80, z-score = -1.15, R) >droVir3.scaffold_12855 8587159 114 + 10161210 GACUUAAUAAGCUCCCAGGUGUACAAUUUUGAGCGAGUCGCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAACAAUUCCACCCGCCCGCACGCCUCCUCG----- ..........((((((..(((((..((..(.((((((((....)))))(((((....))))).))).).))..)))))..)))).............(....)..)).......----- ( -27.80, z-score = -0.82, R) >droMoj3.scaffold_6540 23783075 96 + 34148556 GACUUAAUAAGCUCCCAGGUGUACAAUUUUCAGCAGGUCGCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAC---U-CUCUCACUG------------------- .........((.((((..(((((..((..(((((.((((....)))).(((((....))))).))))).))..)))))..)))).)---)-.........------------------- ( -27.80, z-score = -2.62, R) >droGri2.scaffold_14830 2281588 105 + 6267026 GACUUAAUAAGCUCCCAGGUGUACGAUUUUCAGCG-----CUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACUGGGGAA----CUCCGCCCUCUCGCCCGCCCCAACA----- ..........((((((.((((((.(((..(((((.-----........(((((....))))).))))).))).)))))).)))).----....((......))..)).......----- ( -26.40, z-score = -0.96, R) >consensus UGCUUAAUAAGCUCCCUGGUGUACAAUUUUCAGC_____GCUCGACUCGUAAUGGAAAUUAUAGCUGAUGUCAUACACAGGGGAAA___UGCCCCGACUGGG__CGCC___CC______ ............((((..(((((..((..(((((.......((.(.......).)).......))))).))..)))))..))))................................... (-18.17 = -18.18 + 0.01)


| Location | 4,909,104 – 4,909,210 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Shannon entropy | 0.41196 |
| G+C content | 0.47567 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.69 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
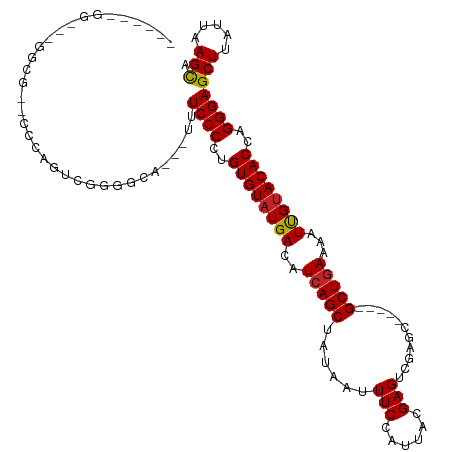
>dm3.chr3R 4909104 106 - 27905053 -----CGGUAAGGCGACCCCAGUCGGAGCA---UUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA -----.((((((.((((....)))).....---.(((((..(((((..(..(((((......(((......)))......-----)))))...)..)))))..)))))))))))..... ( -28.50, z-score = -1.15, R) >droSim1.chr3R 11111430 106 + 27517382 -----CGGUAAGGCGACCCCAGUCGGAGCA---UUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA -----.((((((.((((....)))).....---.(((((..(((((..(..(((((......(((......)))......-----)))))...)..)))))..)))))))))))..... ( -28.50, z-score = -1.15, R) >droSec1.super_0 4148870 85 + 21120651 --------------------------AGCA---UUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA --------------------------....---....(((((((((..(..(((((......(((......)))......-----)))))...)..)))).))))).(((.....))). ( -22.90, z-score = -2.46, R) >droYak2.chr3R 8955645 105 - 28832112 -----UGGUGGGGCG-UCCCACUCGGGGCA---GCUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGCACACCAGGGAGCUUAUUAAGCA -----.......(((-((((....)))))(---((((((.((((((.....(((((......(((......)))......-----)))))....))))))...)))))))......)). ( -36.10, z-score = -1.39, R) >droEre2.scaffold_4770 1053008 106 + 17746568 -----UGGCAGGGCGAACCCACUCGGGGCA---UUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA -----..((.(((....))).....((((.---..((((..(((((..(..(((((......(((......)))......-----)))))...)..)))))..)))))))).....)). ( -29.50, z-score = -0.52, R) >droAna3.scaffold_13340 9067244 111 + 23697760 UGUAAUGGAAAAGCGACCCGAGUCAGGGCA---UUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA ..........(((((.(((......)))).---....(((((((((..(..(((((......(((......)))......-----)))))...)..)))).))))).))))........ ( -30.00, z-score = -1.45, R) >dp4.chr2 6361989 87 + 30794189 ------------------------GGGCAG---AGUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA ------------------------.....(---((..(((((((((..(..(((((......(((......)))......-----)))))...)..)))).)))))..)))........ ( -25.30, z-score = -1.90, R) >droPer1.super_3 169654 87 + 7375914 ------------------------GGGCAG---AGUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC-----GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA ------------------------.....(---((..(((((((((..(..(((((......(((......)))......-----)))))...)..)))).)))))..)))........ ( -25.30, z-score = -1.90, R) >droVir3.scaffold_12855 8587159 114 - 10161210 -----CGAGGAGGCGUGCGGGCGGGUGGAAUUGUUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGCGACUCGCUCAAAAUUGUACACCUGGGAGCUUAUUAAGUC -----(.(((.(.(((((..(((((.((((....)))))))))))))).)....(.((((((((......((((((....))))))....))))))))).))).).............. ( -31.60, z-score = 0.09, R) >droMoj3.scaffold_6540 23783075 96 - 34148556 -------------------CAGUGAGAG-A---GUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGCGACCUGCUGAAAAUUGUACACCUGGGAGCUUAUUAAGUC -------------------........(-(---((((((..(((((..(..(((((.......((......))(((....)))..)))))...)..)))))..))))))))........ ( -28.20, z-score = -2.77, R) >droGri2.scaffold_14830 2281588 105 - 6267026 -----UGUUGGGGCGGGCGAGAGGGCGGAG----UUCCCCAGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAG-----CGCUGAAAAUCGUACACCUGGGAGCUUAUUAAGUC -----......(((..((......)).(((----(((((..((((((((..(((((......(((......))).....-----.)))))...))))))))..)))))))).....))) ( -32.60, z-score = -0.87, R) >consensus ______GG___GGCG__CCCAGUCGGGGCA___UUUCCCCUGUGUAUGACAUCAGCUAUAAUUUCCAUUACGAGUCGAGC_____GCUGAAAAUUGUACACCAGGGAGCUUAUUAAGCA ...................................((((..((((((((..(((((......(((.((.....)).)))......)))))...))))))))..))))(((.....))). (-20.79 = -20.69 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:54 2011