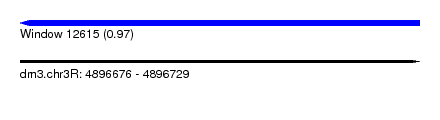
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,896,676 – 4,896,729 |
| Length | 53 |
| Max. P | 0.968038 |

| Location | 4,896,676 – 4,896,729 |
|---|---|
| Length | 53 |
| Sequences | 10 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.50233 |
| G+C content | 0.64525 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -13.68 |
| Energy contribution | -12.81 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
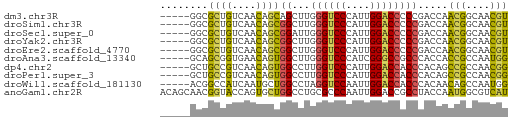
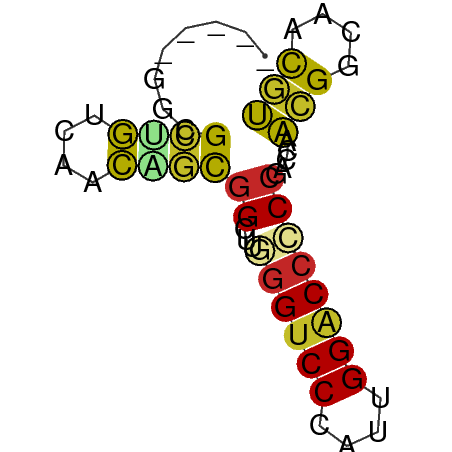
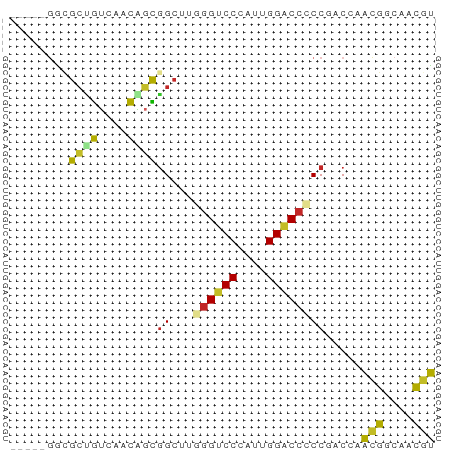
>dm3.chr3R 4896676 53 - 27905053 -----GGCGCUGUCAACAGCAGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGU -----((((((((.....)))))..((((((....))))))..).)).(((....))) ( -22.40, z-score = -2.29, R) >droSim1.chr3R 11099177 53 + 27517382 -----GGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGU -----(.(((((....))))).)..((((((....)))))).......(((....))) ( -23.80, z-score = -2.13, R) >droSec1.super_0 4136724 53 + 21120651 -----GGCGCUGUCAACAGCGGAUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGU -----(((((((....))))((...((((((....))))))))).)).(((....))) ( -23.40, z-score = -2.26, R) >droYak2.chr3R 8943545 53 - 28832112 -----GGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGU -----(.(((((....))))).)..((((((....)))))).......(((....))) ( -23.80, z-score = -2.13, R) >droEre2.scaffold_4770 1041174 53 + 17746568 -----GGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGU -----(.(((((....))))).)..((((((....)))))).......(((....))) ( -23.80, z-score = -2.13, R) >droAna3.scaffold_13340 9055312 53 + 23697760 -----GCAGCGGUGAACAGUGGCUUGGGUCCCAUCGGGCCGCCCACCACCGCCAAUGG -----...((((((....(((((((((......)))))))))....))))))...... ( -23.70, z-score = -1.19, R) >dp4.chr2 6350209 53 + 30794189 -----GCUGCCGUCAACAGUGGCCUUGGUCCCAUUGGACCACCCACAGCCGCCAACGG -----....((((.....(((((..((((((....))))))......)))))..)))) ( -17.80, z-score = -1.04, R) >droPer1.super_3 157528 53 + 7375914 -----GCUGCCGUCAACAGUGGCCUUGGUCCCAUUGGACCACCCACAGCCGCCAACGG -----....((((.....(((((..((((((....))))))......)))))..)))) ( -17.80, z-score = -1.04, R) >droWil1.scaffold_181130 8257205 53 + 16660200 -----ACGGCCAUCAAUGCUGGCCUAGGUCCAAUUGGACCACCCACAACAGCCAAUGG -----..(((((.......)))))..(((((....))))).................. ( -17.30, z-score = -1.70, R) >anoGam1.chr2R 47904049 58 - 62725911 ACAGCAACGGUACCAGUGCUGGCCUGCGCCCAAUUGGACCGCCUACCAAUGGCGUCAU .(((((..(....)..)))))....(((((..(((((........))))))))))... ( -18.70, z-score = -0.54, R) >consensus _____GGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGU ........((((....))))((...((((((....)))))))).....(((....))) (-13.68 = -12.81 + -0.87)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:51 2011