
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,869,540 – 4,869,633 |
| Length | 93 |
| Max. P | 0.590728 |

| Location | 4,869,540 – 4,869,633 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 64.84 |
| Shannon entropy | 0.68980 |
| G+C content | 0.46592 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -10.19 |
| Energy contribution | -9.26 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
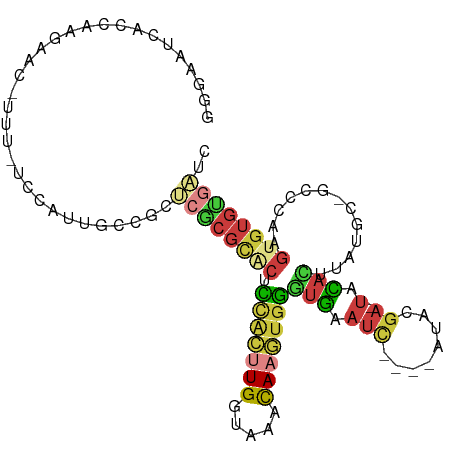
>dm3.chr3R 4869540 93 - 27905053 GGGAAUCACUAA-----UUU-UUCAUUGCCGCUCGCGCACGCCGCUUGGUAAACAAGUGGGUGAAUC----AUAC-A-UGUACUUAUGC-GCCUAAGUGUGUGUUG ..((.((((...-----...-.....((((....).)))..(((((((.....))))))))))).))----..((-(-..((((((...-...))))))..))).. ( -26.50, z-score = -0.79, R) >droAna3.scaffold_13340 9012710 103 + 23697760 GGGAUUCACUCGGUUUGUUUAUGCAUCGUCCCGAUCGCCCCUC-CCUGGCAAAUAUUCUGGUGAAUCACCGAUCCGA-UACAUAUAUUC-GCCCAAGUUUGUGAUC (((((((((.(((..(((((..(.((((...)))))(((....-...))))))))..)))))))))).))(((((((-.((........-......))))).)))) ( -25.14, z-score = -0.54, R) >droEre2.scaffold_4770 1013724 98 + 17746568 GGGAACCACAUAGAAU-UUU-UCCACUGCCGCUCGCGCGCUUCACUUGGUAAACAAGUGGGUGACUC----ACACGC-UACCCUUAUGC-GUCCAAGUGUGUGAUC .(..(((.........-...-......((((....)).))..((((((.....)))))))))..)((----((((((-(((.(....).-))...))))))))).. ( -26.70, z-score = -0.55, R) >droYak2.chr3R 8916174 99 - 28832112 GGGAAUCACCUAGAACACUU-UCCACCGCCCCUCGCGCACUUCACUUGGUAAACAAGUGGGUGAAUC----ACACGA-UACACUUAUGC-GUCCAAGAGUGCGAUC .((((..............)-)))..(((..((((((((...((((((.....))))))((((.(((----....))-).))))..)))-))....))).)))... ( -26.34, z-score = -1.42, R) >droSec1.super_0 4109881 99 + 21120651 GGGAAUCACCAAGAACAUUU-CCCAUUGCCGCUCGCGCACUCCGCUUGGUAAACAAGUGGGUGAAUC----AUACGA-UACACUUAUGC-GCCCAAGUGUGUGAUC (((((.............))-)))((..(((((.(((((..(((((((.....)))))))(((.(((----....))-).)))...)))-))...)))).)..)). ( -34.22, z-score = -3.08, R) >droSim1.chr3R 11071715 99 + 27517382 GGGAAUCACCAAGAACAUUU-UCCAUUGCCGCUCGCGCACUCCGCUUGGUAAACAAGUGGCUGAAUC----AUACGA-UACACUUAUGC-GCCCAAGUGUGUGAUC (((((.............))-)))((..(((((.(((((..(((((((.....))))))).((.(((----....))-).))....)))-))...)))).)..)). ( -27.12, z-score = -1.20, R) >droGri2.scaffold_14906 8865949 101 - 14172833 UAUAAUCACCAG-----CUUAUAAAAUCUACUAAAACUGGAAUAUAUGAAAUAUACAUGUAUAUAUUUUGAGUCCGAGUAUAUACCCACAAUUGAAAUGCAUAUUC .....(((((((-----...................))))..................((((((((((.......)))))))))).......)))........... ( -10.31, z-score = 0.55, R) >consensus GGGAAUCACCAAGAAC_UUU_UCCAUUGCCGCUCGCGCACUCCACUUGGUAAACAAGUGGGUGAAUC____AUACGA_UACACUUAUGC_GCCCAAGUGUGUGAUC (((.......................(((.....)))(((.(((((((.....))))))))))............................)))............ (-10.19 = -9.26 + -0.93)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:45 2011