
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,836,541 – 4,836,609 |
| Length | 68 |
| Max. P | 0.500000 |

| Location | 4,836,541 – 4,836,609 |
|---|---|
| Length | 68 |
| Sequences | 12 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 65.31 |
| Shannon entropy | 0.70447 |
| G+C content | 0.63936 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -9.45 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
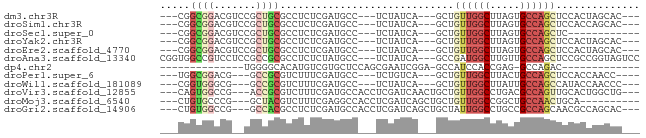
>dm3.chr3R 4836541 68 + 27905053 ---CGGCGGACGUCCGCUGCGCCUCUCGAUGCC---UCUAUCA---GCUGUUGGCUUAGUGCCAGCUCCACUAGCAC--- ---((((((....)))))).((.....((((..---..)))).---(..((((((.....))))))..)....))..--- ( -22.30, z-score = -0.38, R) >droSim1.chr3R 11038896 68 - 27517382 ---CGGCGGACGUCCGCUGCGCCUCUCGAUGCC---UCUAUCA---GCUGUUGGCUUAGUGCCAGCUCCACCAGCAC--- ---((((((....)))))).((.....((((..---..)))).---(..((((((.....))))))..)....))..--- ( -22.30, z-score = -0.34, R) >droSec1.super_0 4077462 59 - 21120651 ---CGGCGGACGUCCGCUGCGCCUCUCGAUGCC---UCUAUCA---GCUGUUGGCUUAGUGCCAGCUC------------ ---((((((....))))))........((((..---..)))).---...((((((.....))))))..------------ ( -20.10, z-score = -0.45, R) >droYak2.chr3R 8883012 68 + 28832112 ---CGGCGGACGUCCGCUGCGCCUCUCGAUGCC---UCUAUCA---GCUGUUGGCUUAGUGCCAGCUCCACUAGCAC--- ---((((((....)))))).((.....((((..---..)))).---(..((((((.....))))))..)....))..--- ( -22.30, z-score = -0.38, R) >droEre2.scaffold_4770 980743 68 - 17746568 ---CGGCGGACGUCCGCUGCGCCUCUCGAUGCC---UCUAUCA---GCUGUUGGCUUAGUGCCAGCUCCACUAGCAC--- ---((((((....)))))).((.....((((..---..)))).---(..((((((.....))))))..)....))..--- ( -22.30, z-score = -0.38, R) >droAna3.scaffold_13340 8977892 74 - 23697760 CGGUGGCCGUCCUCCGCCGCGCCUCUCUAUGCC---UCUAUCA---GCCGAUGGCUUGUUGCCAGCUCCGCCGGUAGUCC ((((((.......))))))..............---.(((((.---((.((((((.....))))..)).)).)))))... ( -21.30, z-score = 0.42, R) >dp4.chr2 27565505 51 + 30794189 --------------UGGGGCACAUGUCGUGCUCCAGCGAAUCGGA-GCCAUCCACCGAG-GCCAGAC------------- --------------((((((((.....))))))))((...((((.-........)))).-)).....------------- ( -17.80, z-score = -0.97, R) >droPer1.super_6 3504603 64 - 6141320 ---UGGCGGACG---GCCGCGUCUUUCGAUGCC---UCUGUCA---GCUGUUGGCUUACUGCCAGCUCCACCAACC---- ---.(((.((((---(..(((((....))))).---.))))).---)))((((((.....))))))..........---- ( -26.10, z-score = -2.70, R) >droWil1.scaffold_181089 2255942 65 + 12369635 ---CGGUGGGCG---GCCGCGUCUUUCGAUGCC---UCUAUCA---GCUGUUGGCUUAUUGCCAGCCAUACCAACCC--- ---.((((((((---((.(((((....))))).---......(---((.....)))....))).))).)))).....--- ( -22.10, z-score = -1.15, R) >droVir3.scaffold_12855 4841295 71 - 10161210 ---CAGUGGCCG---ACCGCGUCUUUCGAUGCCACCUCGAUCAACUGCUGUUGGCCUGACGCCAGUUGCACUGGCUG--- ---....(((((---((.((((...((((.......))))...)).)).)))))))....(((((.....)))))..--- ( -27.50, z-score = -2.40, R) >droMoj3.scaffold_6540 11565030 64 - 34148556 ---CUGUGCCCG---GCUACGUCUUUCGAGGCCACCUCGAUCAGCUGCUGUUGGCCGGCUGCCAACUGCA---------- ---.((.(((((---((........((((((...)))))).((((....)))))))))..))))......---------- ( -21.80, z-score = -0.42, R) >droGri2.scaffold_14906 9186096 71 + 14172833 ---CUGUGGCCG---GCCACGCCUCUCGAUGCCACCUCGAUCAGCUGCUAUUGGCCUGCCGCCAGCAACGCCAGCAC--- ---..(((((.(---((((.((((.((((.......))))..))..))...))))).)))))..((.......))..--- ( -21.50, z-score = 0.20, R) >consensus ___CGGCGGACG__CGCUGCGCCUCUCGAUGCC___UCUAUCA___GCUGUUGGCUUAGUGCCAGCUCCACCAGCAC___ .....((((.......)))).............................((((((.....)))))).............. ( -9.45 = -9.83 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:42 2011