
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,783,522 – 6,783,652 |
| Length | 130 |
| Max. P | 0.606166 |

| Location | 6,783,522 – 6,783,615 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 63.21 |
| Shannon entropy | 0.69826 |
| G+C content | 0.43154 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.05 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
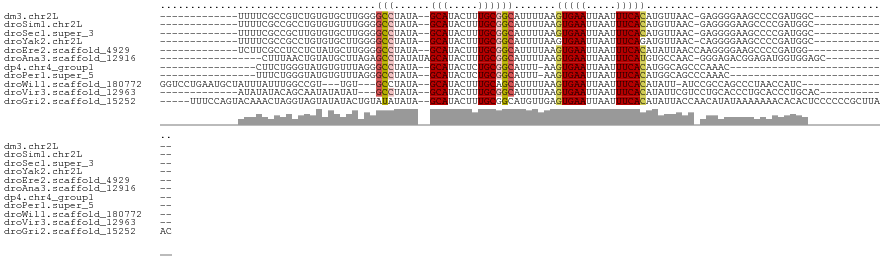
>dm3.chr2L 6783522 93 + 23011544 -------------UUUUCGCCGUCUGUGUGCUUGGGGCCUAUA--GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAAC-GAGGGGAAGCCCCGAUGGC------------- -------------.....(((((..(((((((.(....)...)--))))))...))))).......(((((.....)))))..((((.(-(.(((....))))).))))------------- ( -32.50, z-score = -2.52, R) >droSim1.chr2L 6584173 93 + 22036055 -------------UUUUCGCCGCCUGUGUGUUUGGGGCCUAUA--GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAAC-GAGGGGAAGCCCCGAUGGC------------- -------------.....(((((..(((((((.(....)...)--))))))...))))).......(((((.....)))))..((((.(-(.(((....))))).))))------------- ( -31.70, z-score = -2.16, R) >droSec1.super_3 2322311 93 + 7220098 -------------UUUUCGCCGCUUGUGUGCUUGGGGCCUAUA--GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAAC-GAGGGGAAGCCCCGAUGGC------------- -------------.....(((((..(((((((.(....)...)--))))))...))))).......(((((.....)))))..((((.(-(.(((....))))).))))------------- ( -34.80, z-score = -3.14, R) >droYak2.chr2L 16199054 93 - 22324452 -------------UUUUCGCCGCCUGUGUGCUUGGGGCCUAUA--GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCAGAUGUUAAC-CAGGGGAAGCCCCGAUGGC------------- -------------.....(((((..(((((((.(....)...)--))))))...)))))........((((.....))))........(-((((((...))))..))).------------- ( -29.00, z-score = -0.86, R) >droEre2.scaffold_4929 15700050 93 + 26641161 -------------UCUUCGCCUCCUCUAUGCUUGGGGCCUAUA--GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUAUUAACCAAGGGGAAGCCCCGAUGG-------------- -------------.....(((.((((.......))))......--(((.....)))))).......(((((.....))))).......(((.((((...))))..)))-------------- ( -25.30, z-score = -1.03, R) >droAna3.scaffold_12916 8362222 93 + 16180835 -----------------CUUUAACUGUAUGCUUAGAGCCUAUAUAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCAUGUGCCAAC-GGGAGACGGAGAUGGUGGAGC----------- -----------------........(((((((............)))))))((..((((((.....(((((.....))))))))))..(-.(....).).....)..))..----------- ( -22.50, z-score = -0.84, R) >dp4.chr4_group1 3890208 76 + 5278887 ----------------CUUCUGGGUAUGUGUUUAGGGCCUAUA--GCAUACUCUGCGGCAUUU-AAGUGAAUUAAUUUCACAUGGCAGCCCAAAC--------------------------- ----------------.....((((((((...(((...)))..--))))))))((.(((..((-(.(((((.....))))).)))..)))))...--------------------------- ( -19.70, z-score = -0.87, R) >droPer1.super_5 5433531 76 - 6813705 ----------------UUUCUGGGUAUGUGUUUAGGGCCUAUA--GCAUACUCUGCGGCAUUU-AAGUGAAUUAAUUUCACAUGGCAGCCCAAAC--------------------------- ----------------.....((((((((...(((...)))..--))))))))((.(((..((-(.(((((.....))))).)))..)))))...--------------------------- ( -19.70, z-score = -0.98, R) >droWil1.scaffold_180772 7413413 98 + 8906247 GGUCCUGAAUGCUAUUUAUUUGGCCGU---UGU---GCCUAUA--GCAUACUUUGCAGCAUUUUAAGUGAAUUAAUUUCACAUAUU-AUCCGCCAGCCCUAACCAUC--------------- (((...(((((((........(((...---...---)))....--(((.....))))))))))...(((((.....))))).....-....))).............--------------- ( -20.60, z-score = -1.40, R) >droVir3.scaffold_12963 16346583 92 - 20206255 -------------AUAUAUACAGCAAUAUAUAU---GCCUAUA--GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUAUUCGUCCUGCACCCUGCACCCUGCAC------------ -------------.........((((....(((---((.....--)))))..)))).(((......(((((.....)))))..........(((.....)))...)))..------------ ( -17.50, z-score = -1.44, R) >droGri2.scaffold_15252 1536071 115 + 17193109 -----UUUCCAGUACAAACUAGGUAGUAUAUACUGUAUAUAUA--GCAUACUUUGCGGCAUGUUGAGUGAAUUAAUUUCACAUAUUACCAACAUAUAAAAAAACACACUCCCCCCGCUUAAC -----................((((((((............((--((((.((....)).)))))).(((((.....)))))))))))))................................. ( -17.40, z-score = -0.42, R) >consensus _____________UUUUCGCCGGCUGUGUGUUUGGGGCCUAUA__GCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAAC_CAGGGGAAGCCCCGAUGG______________ ....................................(((......(((.....)))))).......(((((.....)))))......................................... ( -7.85 = -8.05 + 0.21)

| Location | 6,783,550 – 6,783,652 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.14 |
| Shannon entropy | 0.56689 |
| G+C content | 0.43303 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.606166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
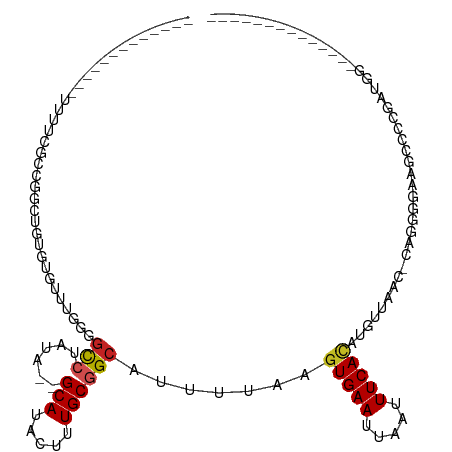
>dm3.chr2L 6783550 102 + 23011544 UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAACGAGGG--GAAGCCCCGAUGGCCCCGCCCACG-----------GCAAACAUU---GCAAGCGUUUAGUUUGCU .((((.(((.((((((.......(((((.....)))))..((((.((.(((--....))))).))))...)))....-----------(((.....)---))..)))...))).)))) ( -31.10, z-score = -1.65, R) >droSim1.chr2L 6584201 102 + 22036055 UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAACGAGGG-GAAGCCCCGAUGGCCC-CGCCCACG-----------GCAAACAUU---GCAAGCGUUUAGUUUGCU .((((.(((.((((((.......(((((.....)))))..((((.((.(((-....))))).))))..-.)))....-----------(((.....)---))..)))...))).)))) ( -31.10, z-score = -1.65, R) >droSec1.super_3 2322339 102 + 7220098 UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAACGAGGG--GAAGCCCCGAUGGCCCCGCCCACG-----------GCAAACAUU---GCAAGCGUUUAGUUUGCU .((((.(((.((((((.......(((((.....)))))..((((.((.(((--....))))).))))...)))....-----------(((.....)---))..)))...))).)))) ( -31.10, z-score = -1.65, R) >droYak2.chr2L 16199082 102 - 22324452 UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCAGAUGUUAACCAGGG--GAAGCCCCGAUGGCCCCGCCCACG-----------GCAAACAUU---GCAAGCGUUUAGUUUGCU .((((.(((..(((((........((((.....))))((((((..((..((--(..(((.....)))....)))..)-----------)..))))))---))...)))..))).)))) ( -28.10, z-score = -0.68, R) >droEre2.scaffold_4929 15700078 102 + 26641161 UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUAUUAACCAAGG--GGAAGCCCCGAUGGCCCGCCCACG-----------GCAAACAUU---GCAAACGUUUAGUUUGCU .((((.(((..(((((.......(((((.....))))).......(((.((--((...))))..)))...)))....-----------(((.....)---))....))..))).)))) ( -29.70, z-score = -2.07, R) >droAna3.scaffold_12916 8362248 104 + 16180835 UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCAUGUGCCAACGGGAGACGGAGAUGGUGGAGCCCUGCCCUUG-----------GCAAAUAUU---AAAAACGUUUACUUUGCU .((((......(((...(((((((((((.....))))).(((((((.(....).)....((..(....)..)).)))-----------)))....))---)))).)))......)))) ( -25.90, z-score = -0.45, R) >dp4.chr4_group1 3890233 83 + 5278887 UAGCAUACUCUGCGGCAUUU-AAGUGAAUUAAUUUCACAUGGCAGCC-----CAAACCCAAACC-------------------------CAAACAGU----CAAACGUUUAGUUUGCU .((((.(((..(((......-..(((((.....))))).(((....)-----))..........-------------------------........----....)))..))).)))) ( -16.70, z-score = -1.56, R) >droPer1.super_5 5433556 83 - 6813705 UAGCAUACUCUGCGGCAUUU-AAGUGAAUUAAUUUCACAUGGCAGCC-----CAAACCCAAACC-------------------------CAAACAGU----CAAACGUUUAGUUUGCU .((((.(((..(((......-..(((((.....))))).(((....)-----))..........-------------------------........----....)))..))).)))) ( -16.70, z-score = -1.56, R) >droWil1.scaffold_180772 7413448 103 + 8906247 UAGCAUACUUUGCAGCAUUUUAAGUGAAUUAAUUUCACAUAUUAUCCG----CCAGCCCUAACCAUCCAUGAUUAAG-----------GCAAACAUUUGGACAAUCGUUUAGUUUGCU .((((.(((..((..........(((((.....)))))......((((----...(((.(((.((....)).))).)-----------)).......)))).....))..))).)))) ( -19.60, z-score = -1.63, R) >droGri2.scaffold_15252 1536107 116 + 17193109 UAGCAUACUUUGCGGCAUGUUGAGUGAAUUAAUUUCACAUAUUACCAACAUAUAAAAAAACACACUCCCCCCGCUUAACCCUCUCAAUGCGAACAUU--ACGGCCCGUUUAGUUAGCU .(((..(((..(((((((((((.(((((.....))))).......))))))....................(((..............)))......--...).))))..)))..))) ( -20.14, z-score = -1.43, R) >consensus UAGCAUACUUUGCGGCAUUUUAAGUGAAUUAAUUUCACAUGUUAACCAGGG__GAACCCCCACCGGCCCCGCCCACG___________GCAAACAUU___GCAAACGUUUAGUUUGCU .((((.(((..(((.........(((((.....)))))..........................((.....))................................)))..))).)))) (-10.43 = -10.97 + 0.54)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:02 2011