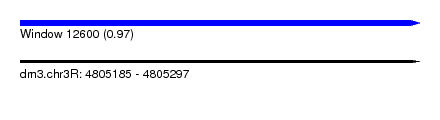
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,805,185 – 4,805,297 |
| Length | 112 |
| Max. P | 0.972747 |

| Location | 4,805,185 – 4,805,297 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.49524 |
| G+C content | 0.49984 |
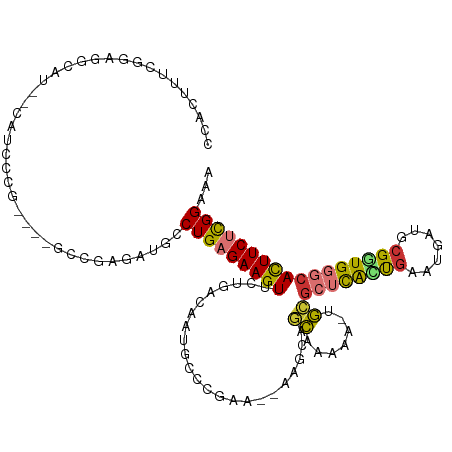
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -16.15 |
| Energy contribution | -15.89 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
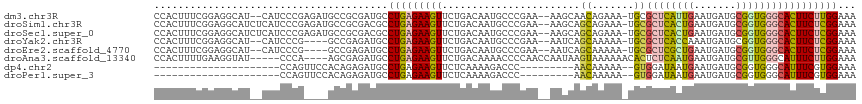
>dm3.chr3R 4805185 112 + 27905053 CCACUUUCGGAGGCAU--CAUCCCGAGAUGCCGCGAUGCCUGAGAAGUUCUGACAAUGCCCGAA--AAGCAACAGAAA-UGCGCUCAUUGAAUGAUGCGGUGGGCACUUCUUGGAAA ......(((..(((((--(.......)))))).)))...(..((((((((((....(((.....--..))).))))..-...((((((((.......))))))))))))))..)... ( -35.60, z-score = -1.19, R) >droSim1.chr3R 11004236 114 - 27517382 CCACUUUCGGAGGCAUCUCAUCCCGAGAUGCCGCGACGCCUGAGAAGUUCUGACAAUGCCCGAA--AAGCAGCAGAAA-UGCGCUCACUGAAUGAUGCGGUGGGCACUUCUCGGAAA ......(((..((((((((.....)))))))).)))...(((((((((((((....(((.....--..))).))))..-...((((((((.......)))))))))))))))))... ( -47.30, z-score = -3.66, R) >droSec1.super_0 4045951 114 - 21120651 CCACUUUCGGAGGCAUCUCAUCCCGAGAUGCCGCGACGCCUGAGAAGUUCUGACAAUGCCCGAA--AAGCAGCAGAAA-UGCGCUCACUGAAUGAUGCGGUGGGCACUUCUCGGAAA ......(((..((((((((.....)))))))).)))...(((((((((((((....(((.....--..))).))))..-...((((((((.......)))))))))))))))))... ( -47.30, z-score = -3.66, R) >droYak2.chr3R 8851260 108 + 28832112 CCACUUUCGGAGGCAU--CAUCCCG----GCCGAGAUGCCUGAGAAGUUCUGACAAUGCCCGAA--AAUCAGCAAAAA-UGCGCUCACCAAAUGAUGCGGUGGGCACUUCUCGGAAA ((........((((((--(.((...----...)))))))))(((((((................--.....(((....-)))(((((((.........))))))))))))))))... ( -35.90, z-score = -1.68, R) >droEre2.scaffold_4770 948843 108 - 17746568 CCACUUUCGGAGGCAU--CAUCCCG----GCCGAGAUGCCUGAGAAGUUCUGACAAUGCCCGAA--AAUCAGCAAAAA-UGCGCUCGCUGAAUGAUGCGGUGGGCACUUCUCGGAAA ....((((..((((((--(.((...----...)))))))))..))))((((((.(((((((...--.....(((....-)))((.(((........)))))))))).)).)))))). ( -36.50, z-score = -1.34, R) >droAna3.scaffold_13340 3554922 108 + 23697760 CCACUUUUGAAGGUAU-----CCCA----AGCGAGAUGCCUGAGAAGUUCUGACAAAACCCCAACCAAUAAGUAAAAAACACUCUCAAUGAAUGAUGCGUUGGGCAUUUCUUGGAAA ..((((((..((((((-----(.(.----...).))))))).))))))((..(.(((..((((((((....((.....))....(((.....))))).))))))..))).)..)).. ( -24.50, z-score = -1.25, R) >dp4.chr2 21556821 85 + 30794189 ---------------------CCAGUUCCACAGAGAUGCCUGAGAAGUUCUCAAAAGACCC---------AACAAAAA--GUGGAUAAUGAAUGAUGCGGUGGGCAUUUCGUGGAAA ---------------------....((((((.((((((((((((.....))).....((((---------(.((....--((.....))...)).)).)))))))))))))))))). ( -27.70, z-score = -3.28, R) >droPer1.super_3 4340850 85 + 7375914 ---------------------CCAGUUCCACAGAGAUGCCUGAGAAGUUCUCAAAAGACCC---------AACAAAAA--GUGGAUAAUGAAUGAUGCGGUGGGCAUUUCGUGGAAA ---------------------....((((((.((((((((((((.....))).....((((---------(.((....--((.....))...)).)).)))))))))))))))))). ( -27.70, z-score = -3.28, R) >consensus CCACUUUCGGAGGCAU__CAUCCCG____GCCGAGAUGCCUGAGAAGUUCUGACAAUGCCCGAA__AAGCAGCAAAAA_UGCGCUCACUGAAUGAUGCGGUGGGCACUUCUCGGAAA .......................................(((((((((.......................((.......))((((((((.......)))))))))))))))))... (-16.15 = -15.89 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:38 2011