| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,799,966 – 4,800,121 |
| Length | 155 |
| Max. P | 0.984656 |

| Location | 4,799,966 – 4,800,121 |
|---|---|
| Length | 155 |
| Sequences | 5 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Shannon entropy | 0.38176 |
| G+C content | 0.41575 |
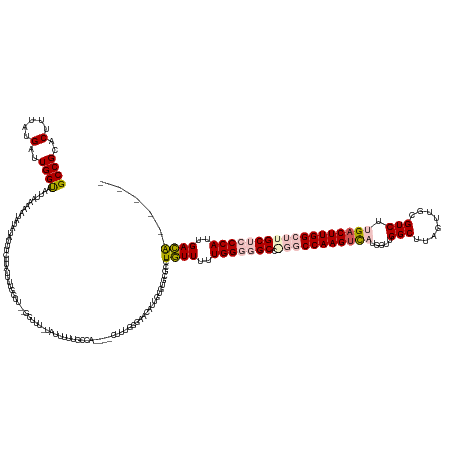
| Mean single sequence MFE | -47.14 |
| Consensus MFE | -27.70 |
| Energy contribution | -29.10 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
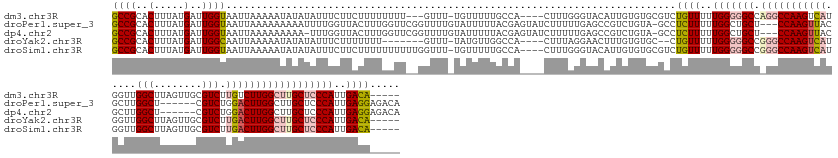
>dm3.chr3R 4799966 155 - 27905053 GCCGCACUUUAUGAUUGGUAAUUAAAAAUAUAUAUUUCUUCUUUUUUUU---GUUU-UGUUUUUGCCA----CUUUGGGUACAUUGUGUGCGUCUGUUUUUGGGGGCCAGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGUCUUGGCUUGCUCCCAUUGACA----- ..(((((.........(((((...((((((..................)---))))-)....)))))(----(..((....))..)))))))...(((..(((((((.((((((((.((.((...((......))..)))).)))))))))))))))..))).----- ( -48.27, z-score = -3.44, R) >droPer1.super_3 4334984 158 - 7375914 GCCGCACUUUAUGAUUGGUAAUUAAAAAAAAAUUUUGGUUACUUUGGUUCGGUUUUGUAUUUUUACGAGUAUCUUUUUGAGCCGUCUGUA-GCCUCUUUUUGGCUGCU---CCAAGUUACGCUUGGCU------CGUCUGGACUUGGCUUGCUCCCAUUGAGGAGACA (((.............(((((((((((.....)))))))))))..(((((((.((((((....)))))).........(((((....(((-(((.......)))))).---..(((.....)))))))------)..))))))).))).((((((......)))).)) ( -45.20, z-score = -2.03, R) >dp4.chr2 21551021 157 - 30794189 GCCGCACUUUAUGAUUGGUAAUUAAAAAAAAA-UUUGGUUACUUUGGUUCGGUUUUGUAUUUUUACGAGUAUCUUUUUGAGCCGUCUGUA-GCCUCUUUUUGGCUGCU---CCAAGUUACGCUUGGCU------CGUCUGGACUUGGCUUGCUCCCAUUGAGGAGACA (((.............((((((((((......-))))))))))..(((((((.((((((....)))))).........(((((....(((-(((.......)))))).---..(((.....)))))))------)..))))))).))).((((((......)))).)) ( -44.50, z-score = -1.81, R) >droYak2.chr3R 8845131 149 - 28832112 GCCGCACUUUAUGAUUGGCAAUUAAAAAUAUAUAUUUCUUUUUUU-------GUUU-UAUGUUGGCCA----CUUUAGGAACUUUGUGUGC--CUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA----- ((((((.(((.(((.(((((((..((((((..............)-------))))-)..))).))))----..))).)))...)))).))--..(((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..))).----- ( -47.54, z-score = -2.54, R) >droSim1.chr3R 10999053 158 + 27517382 GCCGCACUUUAUGAUUGGUAAUUAAAAAUAUAUAUUUCUUCUUUUUUUUUUGGUUU-UGUUUUUGCCA----CUUUGGGUACAUUGUGUGCGUCUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA----- ..(((((...(((......(((((((((..................))))))))).-......((((.----.....)))))))...)))))...(((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..))).----- ( -50.17, z-score = -3.49, R) >consensus GCCGCACUUUAUGAUUGGUAAUUAAAAAUAUAUAUUUCUUAUUUUGGUU__GGUUU_UAUUUUUGCCA____CUUUGGGAACAUUGUGUGCGCCUGUUUUUGGGGGCC_GGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA_____ ((((..(.....)..))))...........................................................................((((..(((((((.(((((((((((.....(((........))).))))))))))))))))))..))))..... (-27.70 = -29.10 + 1.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:37 2011