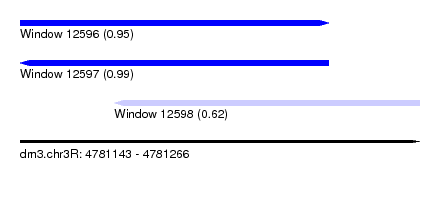
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,781,143 – 4,781,266 |
| Length | 123 |
| Max. P | 0.988109 |

| Location | 4,781,143 – 4,781,238 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.39259 |
| G+C content | 0.46052 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.25 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
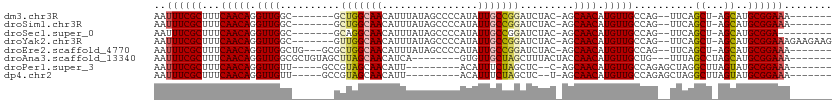
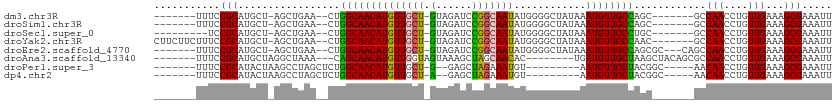
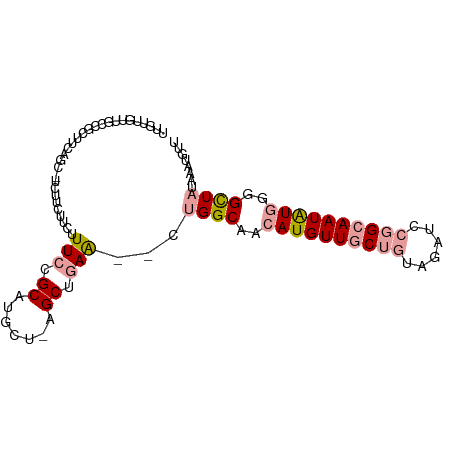
>dm3.chr3R 4781143 95 + 27905053 AAUUUCGCUUUCAACAGGUUGGC-------GCUGGCAACAUUUAUAGCCCCAUAUUGCCGGAUCUAC-AGCAACAUGUUGCCAG--UUCAGCU-AGCAUGCGGAAA------- ..((((((.........((((((-------(((((((((((.(((......)))((((.(......)-.)))).))))))))))--)...)))-)))..)))))).------- ( -30.20, z-score = -1.95, R) >droSim1.chr3R 10979603 95 - 27517382 AAUUUCGCUUUCAACAGGUUGGC-------GCUGGCAACAUUUAUAGCCCCAUAUUGCCGGAUCUAC-AGCAACAUGUUGCCAG--UUCAGCU-AGCAUGCGGAAA------- ..((((((.........((((((-------(((((((((((.(((......)))((((.(......)-.)))).))))))))))--)...)))-)))..)))))).------- ( -30.20, z-score = -1.95, R) >droSec1.super_0 4022049 93 - 21120651 AAUUUCGCUUUCAACAGGUUGGC-------GCAGGCAACAUUUAUAGCCCCAUAUUGCCGGAUCUAC-AGCAACAUGUUGCCAG--UUCAGCU-AGCAUGCGGA--------- ...(((((.........((((((-------((.((((((((.(((......)))((((.(......)-.)))).)))))))).)--)...)))-)))..)))))--------- ( -23.10, z-score = 0.17, R) >droYak2.chr3R 8825471 102 + 28832112 AAUUUCGCUUUCAACAGGUUGGC-------GUUGGCAACAUUUAUAGCCCCAUAUUGCCGGAUCUAC-AGCAACAUGUUGCCAG--UUCAGCU-AGCAUGCGGAAAGAAGAAG ..(((..(((((..((.((((((-------.((((((((((.(((......)))((((.(......)-.)))).))))))))))--....)))-))).))..)))))..))). ( -31.20, z-score = -2.24, R) >droEre2.scaffold_4770 924148 99 - 17746568 AAUUUCGCUUUCAACAGGUUGGCUG---GCGCUGGCAACAUUUAUAGCCCCAUAUUGCCGGAUCUAC-AGCAACAUGUUGCCAG--UUCAGCU-AGCAUGCGGAAA------- ..((((((.........((((((((---(.(((((((((((.(((......)))((((.(......)-.)))).))))))))))--)))))))-)))..)))))).------- ( -36.30, z-score = -2.83, R) >droAna3.scaffold_13340 3531077 95 + 23697760 AAUUUCGCUUUCAACAGGUUGGCGCUGUAGCUUAGCAACAUCA--------GUGUUGCUAGCUUUACUACCAACAUGUUGCUG---UUUAGCCUAGCAUGCGGAAA------- ..((((((...(((((.(((((......((((.(((((((...--------.)))))))))))......))))).))))).((---((......)))).)))))).------- ( -32.90, z-score = -2.46, R) >droPer1.super_3 4311004 89 + 7375914 AAUUUCGCUUUCAACAGGUUGUU-----GCCGUAGCAACAUU---------ACAUUUCUAGCUC--C-AGCAACAUGUUGCCAGAGCUAGGCUUAGUAUGCGGAAA------- ..((((((...(....(..((((-----((....))))))..---------.)...((((((((--.-.((((....))))..))))))))....)...)))))).------- ( -29.90, z-score = -3.28, R) >dp4.chr2 21527056 89 + 30794189 AAUUUCGCUUUCAACAGGUUGUU-----GCCGUAGCAACAUU---------ACAUUUCUAGCUC--U-AGCAACAUGUUGCCAGAGCUAGGCUUAGUAUGCGGAAA------- ..((((((...(....(..((((-----((....))))))..---------.)...((((((((--(-.((((....)))).)))))))))....)...)))))).------- ( -32.00, z-score = -3.92, R) >consensus AAUUUCGCUUUCAACAGGUUGGC_______GCUGGCAACAUUUAUAGCCCCAUAUUGCCGGAUCUAC_AGCAACAUGUUGCCAG__UUCAGCU_AGCAUGCGGAAA_______ ..((((((...(((((.((((..........(((((((................))))))).........)))).)))))..........((...))..))))))........ (-17.47 = -17.25 + -0.22)
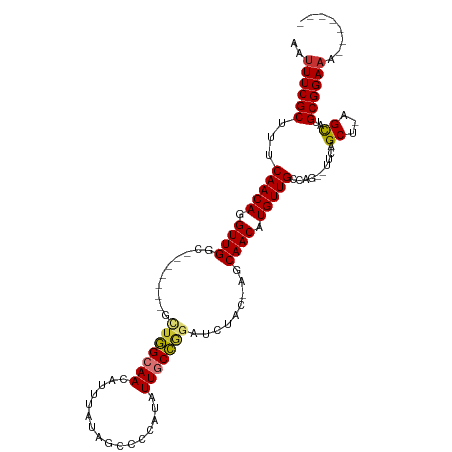
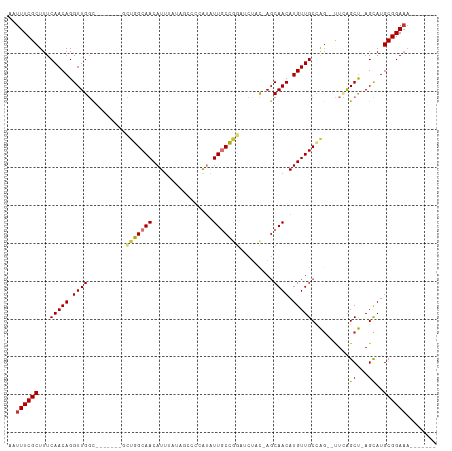
| Location | 4,781,143 – 4,781,238 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.39259 |
| G+C content | 0.46052 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -19.17 |
| Energy contribution | -18.49 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
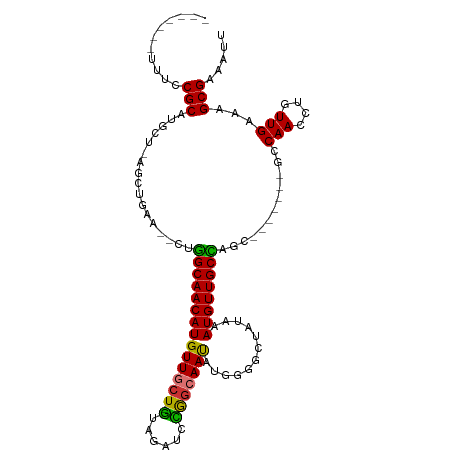
>dm3.chr3R 4781143 95 - 27905053 -------UUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCU-GUAGAUCCGGCAAUAUGGGGCUAUAAAUGUUGCCAGC-------GCCAACCUGUUGAAAGCGAAAUU -------....(((..((.-(((....--.((((..(((((((((-(......))))))))))..)))).....)))))((((-------(......)))))...)))..... ( -29.60, z-score = -1.18, R) >droSim1.chr3R 10979603 95 + 27517382 -------UUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCU-GUAGAUCCGGCAAUAUGGGGCUAUAAAUGUUGCCAGC-------GCCAACCUGUUGAAAGCGAAAUU -------....(((..((.-(((....--.((((..(((((((((-(......))))))))))..)))).....)))))((((-------(......)))))...)))..... ( -29.60, z-score = -1.18, R) >droSec1.super_0 4022049 93 + 21120651 ---------UCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCU-GUAGAUCCGGCAAUAUGGGGCUAUAAAUGUUGCCUGC-------GCCAACCUGUUGAAAGCGAAAUU ---------..((((.((.-(((....--.((((..(((((((((-(......))))))))))..)))).....))))).)))-------).(((....)))........... ( -29.30, z-score = -1.39, R) >droYak2.chr3R 8825471 102 - 28832112 CUUCUUCUUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCU-GUAGAUCCGGCAAUAUGGGGCUAUAAAUGUUGCCAAC-------GCCAACCUGUUGAAAGCGAAAUU .(((..(((((.(((((((-((.....--)))))).(((((((((-(......)))))))))).(((..........)))...-------.......))).))))).)))... ( -30.10, z-score = -1.75, R) >droEre2.scaffold_4770 924148 99 + 17746568 -------UUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCU-GUAGAUCCGGCAAUAUGGGGCUAUAAAUGUUGCCAGCGC---CAGCCAACCUGUUGAAAGCGAAAUU -------....(((.....-.((((..--((((((((((.....(-((((..(((......)))..))))).))))))))))...---))))(((....)))...)))..... ( -33.00, z-score = -1.57, R) >droAna3.scaffold_13340 3531077 95 - 23697760 -------UUUCCGCAUGCUAGGCUAAA---CAGCAACAUGUUGGUAGUAAAGCUAGCAACAC--------UGAUGUUGCUAAGCUACAGCGCCAACCUGUUGAAAGCGAAAUU -------....(((..(((.(......---))))((((.((((((.((..(((((((((((.--------...))))))).))))...)))))))).))))....)))..... ( -31.40, z-score = -2.46, R) >droPer1.super_3 4311004 89 - 7375914 -------UUUCCGCAUACUAAGCCUAGCUCUGGCAACAUGUUGCU-G--GAGCUAGAAAUGU---------AAUGUUGCUACGGC-----AACAACCUGUUGAAAGCGAAAUU -------....(((.(((.....(((((((..((((....)))).-.--)))))))....))---------).((((((....))-----))))...........)))..... ( -36.30, z-score = -5.51, R) >dp4.chr2 21527056 89 - 30794189 -------UUUCCGCAUACUAAGCCUAGCUCUGGCAACAUGUUGCU-A--GAGCUAGAAAUGU---------AAUGUUGCUACGGC-----AACAACCUGUUGAAAGCGAAAUU -------....(((.(((.....(((((((((((((....)))))-)--)))))))....))---------).((((((....))-----))))...........)))..... ( -37.30, z-score = -6.15, R) >consensus _______UUUCCGCAUGCU_AGCUGAA__CUGGCAACAUGUUGCU_GUAGAUCCGGCAAUAUGGGGCUAUAAAUGUUGCCAGC_______GCCAACCUGUUGAAAGCGAAAUU ...........(((.......(((.......)))((((.((((...........((((((((..........))))))))............)))).))))....)))..... (-19.17 = -18.49 + -0.69)
| Location | 4,781,172 – 4,781,266 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 72.28 |
| Shannon entropy | 0.47287 |
| G+C content | 0.45597 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -11.30 |
| Energy contribution | -11.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
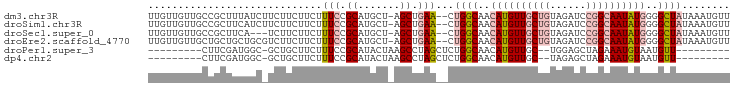
>dm3.chr3R 4781172 94 - 27905053 UUGUUGUUGCCGCUUUAUCUUCUUCUUCUUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCUGUAGAUCCGGCAAUAUGGGGCUAUAAAUGUU ...((((.(((.........................((((-((.....--)))))).((((((((((......)))))))))).))).))))..... ( -26.20, z-score = -1.70, R) >droSim1.chr3R 10979632 94 + 27517382 UUGUUGUUGCCGCUUCAUCUUCUUCUUCUUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCUGUAGAUCCGGCAAUAUGGGGCUAUAAAUGUU ...((((.(((.........................((((-((.....--)))))).((((((((((......)))))))))).))).))))..... ( -26.20, z-score = -1.52, R) >droSec1.super_0 4022078 91 + 21120651 UUGUUGUUGCCGCUUCA---UCUUCUUCUUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCUGUAGAUCCGGCAAUAUGGGGCUAUAAAUGUU ...((((.(((......---................((((-((.....--)))))).((((((((((......)))))))))).))).))))..... ( -26.20, z-score = -1.50, R) >droEre2.scaffold_4770 924181 94 + 17746568 UUGUUGUUGCUGCUGCUGCGUCUUCUUCUUUCCGCAUGCU-AGCUGAA--CUGGCAACAUGUUGCUGUAGAUCCGGCAAUAUGGGGCUAUAAAUGUU .....(((.(.(((((((((............)))).)).-))).)))--)((((..((((((((((......))))))))))..))))........ ( -29.60, z-score = -1.75, R) >droPer1.super_3 4311035 76 - 7375914 ---------CUUCGAUGGC-GCUGCUUCUUUCCGCAUACUAAGCCUAGCUCUGGCAACAUGUUGC--UGGAGCUAGAAAUGUAAUGUU--------- ---------.(((...(((-..(((........)))......)))((((((..((((....))))--..)))))))))..........--------- ( -24.50, z-score = -2.08, R) >dp4.chr2 21527087 76 - 30794189 ---------CUUCGAUGGC-GCUGCUUCUUUCCGCAUACUAAGCCUAGCUCUGGCAACAUGUUGC--UAGAGCUAGAAAUGUAAUGUU--------- ---------.(((...(((-..(((........)))......)))((((((((((((....))))--)))))))))))..........--------- ( -25.50, z-score = -2.58, R) >consensus UUGUUGUUGCCGCUUCAGC_UCUUCUUCUUUCCGCAUGCU_AGCUGAA__CUGGCAACAUGUUGCUGUAGAUCCGGCAAUAUGGGGCUAUAAAUGUU .................................((.......(((.......)))..((((((((((......))))))))))..)).......... (-11.30 = -11.97 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:36 2011