| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,769,176 – 4,769,269 |
| Length | 93 |
| Max. P | 0.545564 |

| Location | 4,769,176 – 4,769,269 |
|---|---|
| Length | 93 |
| Sequences | 14 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.98 |
| Shannon entropy | 0.65403 |
| G+C content | 0.49575 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -9.57 |
| Energy contribution | -8.86 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
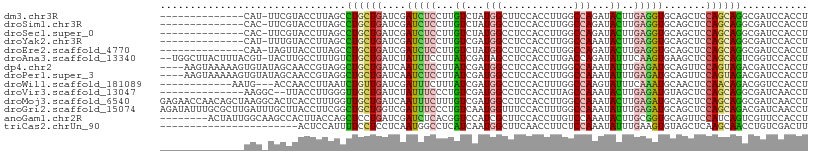
>dm3.chr3R 4769176 93 + 27905053 --------------CAU-UUCGUACCUUAGCCUGCUGAUCGAUCUCCUUGUCUAUGGCUUCCACCUUGGCCAGAUACUUGAGGUGCAGCUCCAGCAGGCGAUCCACCU --------------...-...........((((((((...(((......)))...((((..(((((..(........)..))))).)))).))))))))......... ( -29.00, z-score = -1.77, R) >droSim1.chr3R 10970530 93 - 27517382 --------------CAC-UUCGUACCUUAGCCUGCUGAUCGAUCUCCUUGUCUAUGGCCUCCACCUUGGCCAGAUACUUGAGGUGCAGCUCCAGCAGGCGAUCCACCU --------------...-...........((((((((..(.(((((..(((((..((((........)))))))))...)))))...)...))))))))......... ( -30.20, z-score = -2.00, R) >droSec1.super_0 4010061 93 - 21120651 --------------CAC-UUCGUACCUUAGCCUGCUGAUCGAUCUCCUUGUCUAUGGCCUCCACCUUGGCCAGAUACUUGAGGUGCAGCUCCAGCAGGCGAUCCACCU --------------...-...........((((((((..(.(((((..(((((..((((........)))))))))...)))))...)...))))))))......... ( -30.20, z-score = -2.00, R) >droYak2.chr3R 8813383 93 + 28832112 --------------CAU-UUUGUACCUUAGCCUGCUGAUCGAUCUCCUUGUCGAUGGCCUCCACCUUGGCCAAAUACUUGAGGUGCAGCUCCAGCAGGCGAUCCACCU --------------...-...........((((((((((((((......))))))(((...(((((..(........)..)))))..))).))))))))......... ( -32.30, z-score = -3.13, R) >droEre2.scaffold_4770 912083 93 - 17746568 --------------CAA-UAGUUACCUUAGCCUGCUGAUCGAUCUCCUUGUCUAUGGCCUCCACCUUGGCCAGAUACUUGAGGUGCAGCUCCAGCAGGCGAUCCACCU --------------...-...........((((((((..(.(((((..(((((..((((........)))))))))...)))))...)...))))))))......... ( -30.20, z-score = -1.96, R) >droAna3.scaffold_13340 3519172 105 + 23697760 --UGGCUUACUUUACGU-UACUUGCCUUUGUCUGCUGAUCUAUUUCCUUAUCGAUAGCCUCCACCUUGACCAGAUAUUUCAAGUGAAGCUCCAGCAGUCGGUCCACCU --.(((..((.....))-.....)))...(.(((((((((.((......)).)))(((.((...(((((.........))))).)).))).)))))).)((....)). ( -19.70, z-score = -0.37, R) >dp4.chr2 7805302 104 - 30794189 ----AAGUAAAAAGUGUAUAGCAACCGUAGGCUGCUGAUCAAUCUCCUUAUCGAUGGCCUCCACCUUGGCCAAAUAUUUGAGAUGCAGUUCCAGUAGACGAUCCACCU ----.....................(((..(((((((....(((((..(((...(((((........))))).)))...))))).)))...))))..)))........ ( -23.70, z-score = -0.94, R) >droPer1.super_3 1610245 104 - 7375914 ----AAGUAAAAAGUGUAUAGCAACCGUAGGCUGCUGAUCAAUCUCCUUAUCGAUGGCCUCCACCUUGGCCAAAUAUUUGAGAUGCAGUUCCAGUAGACGAUCCACCU ----.....................(((..(((((((....(((((..(((...(((((........))))).)))...))))).)))...))))..)))........ ( -23.70, z-score = -0.94, R) >droWil1.scaffold_181089 6417665 93 + 12369635 ------------AAUG---ACCAACCUUAAUCUGUUGAUCGAUUUCUUUAUCGAUGGCCUCCACUUUGGCCAAGUAUUUCAAAUGCAACUCCAACAGACGGUCCACCU ------------...(---(((........(((((((((((((......))))))((((........))))..((((.....)))).....))))))).))))..... ( -25.50, z-score = -4.09, R) >droVir3.scaffold_13047 3181183 92 + 19223366 --------------AAGGC--UUACCUUGGGUUGCUGAUCUAUUUCCCUGUCGAUGGCCUCCACCUUAGCCAAAUACUUGAGAUGUAGCUCCAGCAGGCGAUCAACCU --------------((((.--...))))((((((..........(((((((.(.((((..........)))).......(((......)))).))))).)).)))))) ( -19.40, z-score = 1.11, R) >droMoj3.scaffold_6540 20222886 108 + 34148556 GAGAACCAACAGCUAAGGCACUCACCUUUGGUUGCUGAUCAAUUUCUUUGUCGAUGGCCUCCACCUUGGCCAAAUACUUGAGAUGCAGCUCCAGCAGGCGAUCAACCU ..((.......((....))..((.(((((((..((((....(((((...((...(((((........)))))...))..))))).)))).)))).))).))))..... ( -30.10, z-score = -1.00, R) >droGri2.scaffold_15074 7554971 108 + 7742996 AGAUAUUUGCGCUUGAUUUGCUUACCUUCGGCUGCUGGUCGAUUUCCCUGUCAAUGGUUUCCACUUUGGCCAAAUAUUUGAGAUGCAGCUCCAGCAGACGAUCAACCU .(((.(((((((.......))........((((((..((((((......)))..(((((........))))).........)))))))))...)))))..)))..... ( -24.20, z-score = 0.31, R) >anoGam1.chr2R 59222795 100 - 62725911 --------ACUAUUGGCAAGCCACUUACCAGCUCCUGAUCGAUCUCACGGUCCAUCGCUUCCACCUUGUCCAAAUACUUGCGGUGCAGUUCCAUCAGUCGUUCCACCU --------.....(((....)))......(((..(((((.((((....))))((((((.....................)))))).......)))))..)))...... ( -16.60, z-score = 0.25, R) >triCas2.chrUn_90 142366 85 - 150287 -----------------------ACUCCAUUUUCCUCCUCAAUGGCCUCAUCAAUGGCUUCAACCUUCUCCAAAUAUUUGAAGUGUAGCUCAAGCAACCUGUCGACUU -----------------------....................((((........))))..................((((((.((.((....)).)))).))))... ( -9.60, z-score = 0.43, R) >consensus ______________CAU_UACUUACCUUAGGCUGCUGAUCGAUCUCCUUGUCGAUGGCCUCCACCUUGGCCAAAUACUUGAGGUGCAGCUCCAGCAGGCGAUCCACCU ...............................((((((....(((((...((...(((............)))...))..))))).......))))))........... ( -9.57 = -8.86 + -0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:32 2011