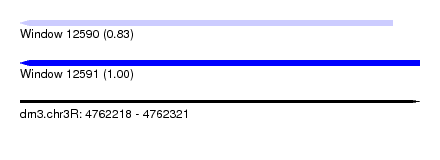
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,762,218 – 4,762,321 |
| Length | 103 |
| Max. P | 0.999588 |

| Location | 4,762,218 – 4,762,314 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 54.71 |
| Shannon entropy | 0.76401 |
| G+C content | 0.54722 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -12.63 |
| Energy contribution | -10.22 |
| Covariance contribution | -2.41 |
| Combinations/Pair | 1.84 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4762218 96 - 27905053 -------CGGUUCGGUUUUCUGGCUUUGGGU---UCU---GCAGCUUGGCUGAUGCCCAGGUGGAGCCAAAUUGAUUGGUUCCUCUGGGGCAUCGGGGCUGUAGAUGUU---------- -------.(((.(((....))))))......---(((---((((((...(((((((((.((.((((((((.....)))))))).)).))))))))))))))))))....---------- ( -46.10, z-score = -4.47, R) >droSim1.chr3R 10963415 96 + 27517382 -------GGGUUCGGGUUUCUUGCUCUGGGU---UCUGCAGCAGCUUGGCUGUUGCCCAGGUGCAGCCAAAUGGGUUGGUUCCUCCGGGGCAUC---GCUGUAGAUGUU---------- -------..(..(((((.....)).)))..)---(((((((((((...)))(.(((((.((.(.((((((.....)))))).).)).))))).)---))))))))....---------- ( -36.40, z-score = -0.43, R) >droSec1.super_0 4003181 96 + 21120651 -------CGGUUCAGUUUUCUUGCUCUGGGU---UCUGCAGCAGCUUGGCUGUUGCCCAGGUGCAGCCAAAUGGGUUGGUUCCUCCGGGGCAUC---GCUGUAGAUGUU---------- -------.....((((.....((((((((..---...((((((((...))))))))..(((..(((((.....)))))...)))))))))))..---))))........---------- ( -35.00, z-score = -0.67, R) >droAna3.scaffold_13340 3512455 85 - 23697760 ---------------------GGUUCUGGCUGGGGUGUGGGUUCUGGCAAUGUUGCUCUGGAUCUGUAACCUGGGUCUGCCGAUCCGGAGUAUC---GACGUAAAUGUU---------- ---------------------.......((..(..(....)..)..)).(((((((((((((((.(((((....)).))).))))))))))...---))))).......---------- ( -29.10, z-score = -1.75, R) >dp4.chr2 7789914 113 + 30794189 GGUUCUUUAGGUUAUCCCGAUGGUUCUGGCU---GUUGGAAUUACGAAGAUGGUUUUCUUGGGCCGGCAACGGCGGUGGCUUCUCUGGAGGAUC---GGCAAAGAUGUUAGCUCGCUGA ((((((((((((((((((((((((....)))---))))).....(.((((......)))).)((((....))))))))))....))))))))))---..........((((....)))) ( -36.80, z-score = -0.55, R) >droPer1.super_3 1596864 113 + 7375914 GGUUCUUUAGGUUAUCCCGAUGGUUCUGGCU---GUUGGAAUUACGAAGAUGGUUUUCUUGGGCCGGCAACGGCGGUGGCUUCUCUGGAGGAUC---GGCAAAGAUGUUAGCUCGCUGA ((((((((((((((((((((((((....)))---))))).....(.((((......)))).)((((....))))))))))....))))))))))---..........((((....)))) ( -36.80, z-score = -0.55, R) >consensus _______CGGUUCAGCUUUCUGGCUCUGGCU___GCUGGAGCAGCGUGGAUGUUGCCCAGGUGCAGCCAAAUGGGUUGGCUCCUCCGGAGCAUC___GCCGUAGAUGUU__________ ............((((....(((((.((((....)))).)))))....)))).(((((.((.(..((((.......))))..).)).)))))........................... (-12.63 = -10.22 + -2.41)

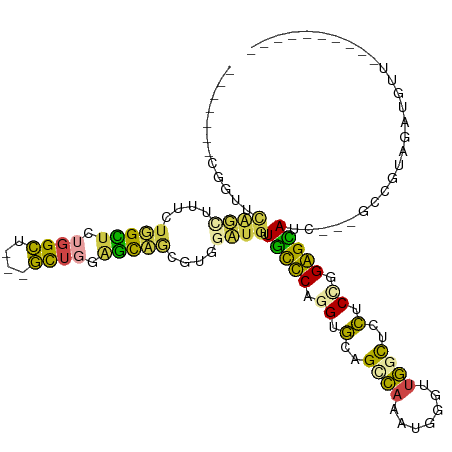
| Location | 4,762,218 – 4,762,321 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 52.49 |
| Shannon entropy | 0.79633 |
| G+C content | 0.52539 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -18.08 |
| Energy contribution | -17.44 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.76 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4762218 103 - 27905053 -------UUUCAGUCGGUUCGGUUUUCUGGCUUUGGGU---UCU---GCAGCUUGGCUGAUGCCCAGGUGGAGCCAAAUUGAUUGGUUCCUCUGGGGCAUCGGGGCUGUAGAUGUU--- -------....((((((.........))))))......---(((---((((((...(((((((((.((.((((((((.....)))))))).)).))))))))))))))))))....--- ( -49.10, z-score = -4.84, R) >droSec1.super_0 4003181 103 + 21120651 -------UUUCAGUCGGUUCAGUUUUCUUGCUCUGGGU---UCUGCAGCAGCUUGGCUGUUGCCCAGGUGCAGCCAAAUGGGUUGGUUCCUCCGGGGCAUC---GCUGUAGAUGUU--- -------.....(((....((((.....((((((((..---...((((((((...))))))))..(((..(((((.....)))))...)))))))))))..---))))..)))...--- ( -37.40, z-score = -1.18, R) >droAna3.scaffold_13340 3512455 88 - 23697760 -------------------------UUUGGUUCUGGCUGGGGUGUGGGUUCUGGCAAUGUUGCUCUGGAUCUGUAACCUGGGUCUGCCGAUCCGGAGUAUC---GACGUAAAUGUU--- -------------------------..........((..(..(....)..)..)).(((((((((((((((.(((((....)).))).))))))))))...---))))).......--- ( -29.10, z-score = -1.75, R) >dp4.chr2 7789921 113 + 30794189 UUCCGUUGGUUCUUUAGGUUAUCCCGAUGGUUCUGGCU---GUUGGAAUUACGAAGAUGGUUUUCUUGGGCCGGCAACGGCGGUGGCUUCUCUGGAGGAUC---GGCAAAGAUGUUAGC ....(((((((((((((((((((((((((((....)))---))))).....(.((((......)))).)((((....))))))))))....))))))))))---)))............ ( -40.60, z-score = -2.06, R) >droPer1.super_3 1596871 113 + 7375914 UUCCGUUGGUUCUUUAGGUUAUCCCGAUGGUUCUGGCU---GUUGGAAUUACGAAGAUGGUUUUCUUGGGCCGGCAACGGCGGUGGCUUCUCUGGAGGAUC---GGCAAAGAUGUUAGC ....(((((((((((((((((((((((((((....)))---))))).....(.((((......)))).)((((....))))))))))....))))))))))---)))............ ( -40.60, z-score = -2.06, R) >consensus _______GGUCAGUCAGGUCAGCCCUAUGGUUCUGGCU___GUUGGAGUUACGAAGAUGGUGCUCUGGGGCAGCCAACUGGGUUGGCUCCUCUGGAGCAUC___GCCGUAGAUGUU___ ....................(((....((((((((((....))))))))))....)))((((((((((((..((((.......))))..)))))))))))).................. (-18.08 = -17.44 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:30 2011