
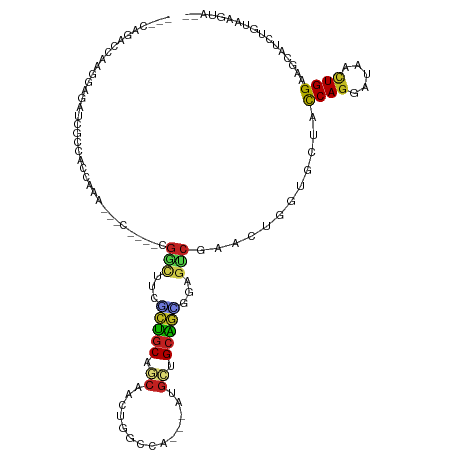
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,761,970 – 4,762,076 |
| Length | 106 |
| Max. P | 0.948891 |

| Location | 4,761,970 – 4,762,075 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.62 |
| Shannon entropy | 0.71884 |
| G+C content | 0.53249 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.50 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.93 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
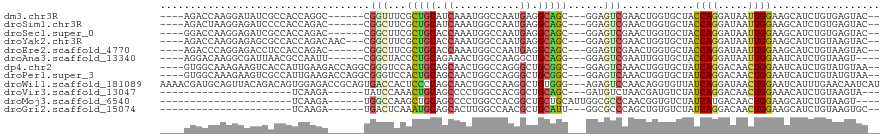
>dm3.chr3R 4761970 105 + 27905053 ---AAGACCAAGGAUAUCGCCACCAGG---C----CGGUUUCGCUGCAUCAAAUGGCCA---AUGAGGCAGCGGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUGAGUA-- ---...((((.((((......(((((.---.----((..((((((((.(((........---.))).))))))))..))..)))))(((.((((.....)))).))))))).)).)).-- ( -40.60, z-score = -2.81, R) >droSim1.chr3R 10963144 105 - 27517382 ---CAGACUAAGGAGAUCCCCACCAGA---C----CGGCUUCGCUGCAUCAAAUGGCCA---AUGAGGCAGCGGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUGAGUA-- ---...(((..(((.......(((((.---.----((((((((((((.(((........---.))).))))))))))))..)))))(((.((((.....)))).))).)))...))).-- ( -42.30, z-score = -3.47, R) >droSec1.super_0 4002910 105 - 21120651 ---AGGACCAAGGAGAUCGCCACCAGA---C----CGGCUUCGCUGCACCAAAUGGCCA---AUGAGGCAGCGGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUGAGUA-- ---...((((.(((.......(((((.---.----((((((((((((..((........---.))..))))))))))))..)))))(((.((((.....)))).))).))).)).)).-- ( -41.30, z-score = -2.78, R) >droYak2.chr3R 8806059 108 + 28832112 ---CAGACCAAGGAGAGCGCCACCAGACAAC----CGGCUUCGCUGCACCAAAUGGCCA---AUGAGGCAGCGGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUAAGUA-- ---((((....((......))(((((.....----((((((((((((..((........---.))..))))))))))))..)))))(((.((((.....)))).))).))))......-- ( -39.20, z-score = -2.52, R) >droEre2.scaffold_4770 904838 105 - 17746568 ---CAGACCCAGGAGACCUCCACCAGA---C----CGGCUUCGCUGCACCAAAUGGCCA---AUGAGGCAGCGGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUAAGUA-- ---((((....(((....)))(((((.---.----((((((((((((..((........---.))..))))))))))))..)))))(((.((((.....)))).))).))))......-- ( -43.80, z-score = -3.89, R) >droAna3.scaffold_13340 3512222 104 + 23697760 ---CAGGACAAGGCGAUUAACGCCAAU---U----CGGCUACCCUGCAGAAACUGGCCA---AGGCUGCAGGGGAGUCGAAUUGGUGCUACCAGGAUAAUUGGAAUCAUCUGUAAGU--- ---....(((.(..((((..(((((((---(----(((((.((((((((...((.....---)).)))))))).)))))))))))))...((((.....))))))))..))))....--- ( -45.80, z-score = -5.13, R) >dp4.chr2 7789575 111 - 30794189 ---GGUGGCAAAGAAGUCACCAUUGAAGACCA-GGCGGGUCCACUGCAGCAACUGGCCA---GGGCUGCGGCGGAGUCAAACUGGUGCUAUCAGGACAACUGGAAUCAUCUGUAUGUA-- ---((((((......))))))..(((((((((-(..((.(((.(((((((..((....)---).))))))).))).))...))))).)).((((.....))))..)))..........-- ( -41.50, z-score = -1.61, R) >droPer1.super_3 1596525 111 - 7375914 ---GGUGGCAAAGAAGUCGCCAUUGAAGACCA-GGCGGGUCCACUGCAGCAACUGGCCA---GGGCUGCGGCGGAGUCAAACUGGUGCUAUCAGGACAACUGGAAUCAUCUGUAUGUA-- ---((((((......))))))..(((((((((-(..((.(((.(((((((..((....)---).))))))).))).))...))))).)).((((.....))))..)))..........-- ( -41.10, z-score = -1.27, R) >droWil1.scaffold_181089 5957872 117 - 12369635 GAAAACGAUGCAGUUACAGACAGUGGAGACCGCAGUGACCACUCCCCAGCAACUGGCCA---AGGCUGUGGGAGAGUCCAACAGGUGUUAUCAGGAUAACUGGAAUCAUUUGAACAAUCA ......(((...(((...(((.((((...))))........(((((((((..((.....---)))))).))))).)))...((((((...((((.....))))...))))))))).))). ( -32.30, z-score = -0.39, R) >droVir3.scaffold_13047 3171214 86 + 19223366 ----------------------UCAAGA-------UAUCCAAACUGCAGCCCCUGGCCA---CGGCUGCAGCGAUGUCUAACGAUGUCUAUCAGGACAACUGGAAACAUCUGUAAGUA-- ----------------------...(((-------((((....((((((((........---.)))))))).)))))))...(((((...((((.....))))..)))))........-- ( -28.70, z-score = -3.10, R) >droMoj3.scaffold_6540 20212563 88 + 34148556 ----------------------UCAAGA-------UGGCCAAGCUGCAGCCCCUGGCCACGGCUGCUGCAUUGGCGCCCAACGGUGUCUAUCAUGACAACUGGAAGCAUCUGUAAGU--- ----------------------...(((-------(((((((((.((((((.........)))))).)).)))))......(((((((......))).))))....)))))......--- ( -33.60, z-score = -1.41, R) >droGri2.scaffold_15074 7544795 86 + 7742996 ----------------------UCAAGA-------UGACUCAAAUGCAGCACUUGGCCA---ACGCUGCAUUGGCGCCCAGCUGUGUCUAUCAGGACAACUGGAAGCAUCUGUAAGUG-- ----------------------...(((-------((.((..((((((((.........---..)))))))).....((((...(((((....))))).)))).))))))).......-- ( -26.00, z-score = -0.64, R) >consensus ___CAGACCAAGGAGAUCGCCACCAAA___C____CGGCUUCGCUGCAGCAACUGGCCA___AUGCUGCAGCGGAGUCGAACUGGUGCUACCAGGAUAACUGGAAGCAUCUGUAAGUA__ ....................................(((...(((((.((..............)).)))))...)))............((((.....))))................. ( -9.80 = -9.50 + -0.30)

| Location | 4,761,971 – 4,762,076 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.68 |
| Shannon entropy | 0.72207 |
| G+C content | 0.53193 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -8.63 |
| Energy contribution | -8.14 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.82 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4761971 105 + 27905053 ----AGACCAAGGAUAUCGCCACCAGGC------CGGUUUCGCUGCAUCAAAUGGCCAAUGAGGCAGC---GGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUGAGUAC-- ----..((((.((((......(((((..------((..((((((((.(((.........))).)))))---)))..))..)))))(((.((((.....)))).))))))).)).))..-- ( -40.60, z-score = -2.90, R) >droSim1.chr3R 10963145 105 - 27517382 ----AGACUAAGGAGAUCCCCACCAGAC------CGGCUUCGCUGCAUCAAAUGGCCAAUGAGGCAGC---GGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUGAGUAC-- ----..(((..(((.......(((((..------((((((((((((.(((.........))).)))))---)))))))..)))))(((.((((.....)))).))).)))...)))..-- ( -42.30, z-score = -3.68, R) >droSec1.super_0 4002911 105 - 21120651 ----GGACCAAGGAGAUCGCCACCAGAC------CGGCUUCGCUGCACCAAAUGGCCAAUGAGGCAGC---GGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUGAGUAC-- ----..((((.(((.......(((((..------((((((((((((..((.........))..)))))---)))))))..)))))(((.((((.....)))).))).))).)).))..-- ( -41.30, z-score = -3.05, R) >droYak2.chr3R 8806060 108 + 28832112 ----AGACCAAGGAGAGCGCCACCAGACAAC---CGGCUUCGCUGCACCAAAUGGCCAAUGAGGCAGC---GGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUAAGUAC-- ----.......(.(((((((.(((((.....---((((((((((((..((.........))..)))))---)))))))..)))))))..((((.....))))..)).))).)......-- ( -38.20, z-score = -2.41, R) >droEre2.scaffold_4770 904839 105 - 17746568 ----AGACCCAGGAGACCUCCACCAGAC------CGGCUUCGCUGCACCAAAUGGCCAAUGAGGCAGC---GGAGUCGAACUGGUGCUACCAGGAUAAUUGGAAGCAUCUGUAAGUAC-- ----.......(((....)))(((((..------((((((((((((..((.........))..)))))---)))))))..)))))(((.((((.....)))).)))............-- ( -41.30, z-score = -3.34, R) >droAna3.scaffold_13340 3512223 103 + 23697760 ----AGGACAAGGCGAUUAACGCCAAUU------CGGCUACCCUGCAGAAACUGGCCAAGGCUGCAGG---GGAGUCGAAUUGGUGCUACCAGGAUAAUUGGAAUCAUCUGUAAGU---- ----...(((.(..((((..((((((((------(((((.((((((((...((.....)).)))))))---).)))))))))))))...((((.....))))))))..))))....---- ( -45.80, z-score = -5.18, R) >dp4.chr2 7789576 111 - 30794189 ----GUGGCAAAGAAGUCACCAUUGAAGACCAGGCGGGUCCACUGCAGCAACUGGCCAGGGCUGCGGC---GGAGUCAAACUGGUGCUAUCAGGACAACUGGAAUCAUCUGUAUGUAA-- ----(((((......)))))...((((((((((..((.(((.(((((((..((....)).))))))).---))).))...))))).)).((((.....))))..)))...........-- ( -37.30, z-score = -0.64, R) >droPer1.super_3 1596526 111 - 7375914 ----GUGGCAAAGAAGUCGCCAUUGAAGACCAGGCGGGUCCACUGCAGCAACUGGCCAGGGCUGCGGC---GGAGUCAAACUGGUGCUAUCAGGACAACUGGAAUCAUCUGUAUGUAA-- ----(((((.........)))))((((((((((..((.(((.(((((((..((....)).))))))).---))).))...))))).)).((((.....))))..)))...........-- ( -37.00, z-score = -0.32, R) >droWil1.scaffold_181089 5957873 117 - 12369635 AAAACGAUGCAGUUACAGACAGUGGAGACCGCAGUGACCACUCCCCAGCAACUGGCCAAGGCUGUGGG---AGAGUCCAACAGGUGUUAUCAGGAUAACUGGAAUCAUUUGAACAAUCAU .....(((...(((...(((.((((...))))........(((((((((..((.....)))))).)))---)).)))...((((((...((((.....))))...))))))))).))).. ( -32.30, z-score = -0.55, R) >droVir3.scaffold_13047 3171214 86 + 19223366 ----------------------UCAAGA------UAUCCAAACUGCAGCCCCUGGCCACGGCUGCAGC---GAUGUCUAACGAUGUCUAUCAGGACAACUGGAAACAUCUGUAAGUA--- ----------------------...(((------((((....((((((((.........)))))))).---)))))))...(((((...((((.....))))..)))))........--- ( -28.70, z-score = -3.10, R) >droMoj3.scaffold_6540 20212563 88 + 34148556 ----------------------UCAAGA------UGGCCAAGCUGCAGCCCCUGGCCACGGCUGCUGCAUUGGCGCCCAACGGUGUCUAUCAUGACAACUGGAAGCAUCUGUAAGU---- ----------------------...(((------(((((((((.((((((.........)))))).)).)))))......(((((((......))).))))....)))))......---- ( -33.60, z-score = -1.41, R) >droGri2.scaffold_15074 7544795 87 + 7742996 ----------------------UCAAGA------UGACUCAAAUGCAGCACUUGGCCAACGCUGCAUU---GGCGCCCAGCUGUGUCUAUCAGGACAACUGGAAGCAUCUGUAAGUGC-- ----------------------...(((------((.((..((((((((...........))))))))---.....((((...(((((....))))).)))).)))))))........-- ( -26.00, z-score = -0.23, R) >consensus ____AGACCAAGGAGAUCGCCACCAAAC______CGGCUUCGCUGCAGCAACUGGCCAAGGCUGCAGC___GGAGUCGAACUGGUGCUACCAGGAUAACUGGAAGCAUCUGUAAGUAC__ .........................................(((((.((...........)).))))).....................((((.....)))).................. ( -8.63 = -8.14 + -0.49)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:29 2011