| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,757,846 – 4,757,958 |
| Length | 112 |
| Max. P | 0.927481 |

| Location | 4,757,846 – 4,757,958 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.15 |
| Shannon entropy | 0.65246 |
| G+C content | 0.38734 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -5.89 |
| Energy contribution | -7.04 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4757846 112 - 27905053 AUCGAUAACUUAUUGAUGAACCAUUUUGUUGUUACUCGCUAGUAAUGGCGAUUCGUUAGCAGUUAAGAGCUUAAAGGCUAGAACGAAAUCCAAAUAGACAGACGAAGUGAAU (((((((...)))))))....((((((((((((..((((((....))))))(((((((((..((((....))))..))))..))))).........)))).))))))))... ( -31.40, z-score = -3.48, R) >droEre2.scaffold_4770 900665 111 + 17746568 UUCGAUAACUUUUCGAUUAAGCAUUUUG-UGUUACUCGCCAGUAGUGGCGAUUAUUUAGCGAUAACCAGCUAGAAGGCCUAUAGCAAAUCAAAAUAGCCCGAUAAAGUGCAU .((((.......))))....((((((((-(.....((((((....))))))...((((((........)))))).((.((((...........)))).)).))))))))).. ( -29.40, z-score = -2.66, R) >droYak2.chr3R 8801923 105 - 28832112 UUCGAUAACUUUUCGAUUAAACAUUUC-UUGUUACUCACCAGUAAUGACGAUUCUUUAGCGGUAACCAGCUAAAAGGCUAUCAACAACUCAAAAUAGCCAGUGCAC------ .((((.......)))).........((-(..(((((....)))))..).))...((((((........)))))).((((((............)))))).......------ ( -19.40, z-score = -1.88, R) >droSec1.super_0 3998692 111 + 21120651 AUCGAUAACUUGUUGAUGAACCAUUUUGUUGUUACUCGCCAGUAAUGGCGAUCCGUUAGCGGUUACCAGC-UAAAGGCUAGAAAGAAAUGUAAAUAGCCAGACGAAGUGAAU ((((((.....))))))....(((((((((.....((((((....))))))....(((((........))-))).(((((...(....).....))))).)))))))))... ( -33.40, z-score = -3.70, R) >droSim1.chr3R 10958925 111 + 27517382 AUCGAUAACUUGUUGAUGAACCAUUUUGUUGUUACUCGCCAGUAAUGGCGAUCCGUUAGCGGUUACCAGC-UAAAGGCUAGAAAGAAAUCUAAAUAGCCAGACGAAGUGAAU ((((((.....))))))....(((((((((.....((((((....))))))....(((((........))-))).(((((...((....))...))))).)))))))))... ( -34.70, z-score = -4.07, R) >droGri2.scaffold_15074 214090 103 + 7742996 CUCUGUAAUUUAAUCAUCAUCAUCAUUAUCAUCAACGGAGACAAGCGACAAAACGACUGAGGCUUUC----UACAAGCCAAUUCCAAUUCUCCACAAAGUACAUGAG----- (((((((...((((.((....)).))))........(((((((..((......))..)).(((((..----...))))).........)))))......)))).)))----- ( -16.90, z-score = -1.67, R) >consensus AUCGAUAACUUAUUGAUGAACCAUUUUGUUGUUACUCGCCAGUAAUGGCGAUUCGUUAGCGGUUACCAGC_UAAAGGCUAGAAACAAAUCUAAAUAGCCAGACGAAGUGAAU ............(((...(((....(((((((((((....)))))))))))...)))..))).............(((((..............)))))............. ( -5.89 = -7.04 + 1.15)

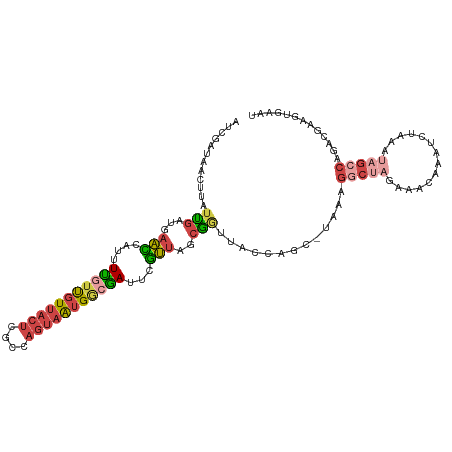
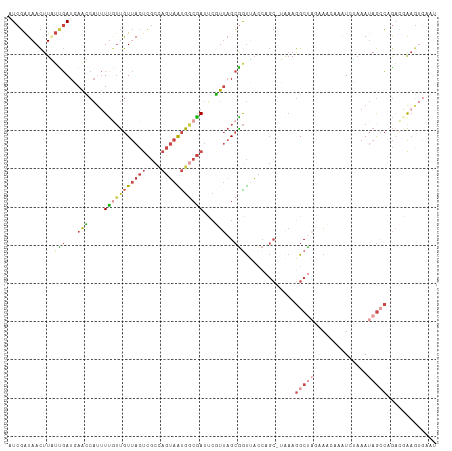
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:27 2011