| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,748,631 – 4,748,730 |
| Length | 99 |
| Max. P | 0.665125 |

| Location | 4,748,631 – 4,748,730 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.79 |
| Shannon entropy | 0.72218 |
| G+C content | 0.46424 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -16.43 |
| Energy contribution | -14.86 |
| Covariance contribution | -1.58 |
| Combinations/Pair | 1.89 |
| Mean z-score | -0.19 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4748631 99 - 27905053 UGUGGAGAU-AUAAUAUUUUUUUACAGUGCUUAGGAUCGGUCUGUAAGCACUUUAUAGAGGG----------GCCACUAUGAUUUUUAUGUGCGCUAGUGCAGGCAGCUU ((((((((.-........))))))))((((((((((....))).)))))))......(((..----------(((..(((((...)))))((((....)))))))..))) ( -22.60, z-score = 0.45, R) >droAna3.scaffold_13340 3499168 97 - 23697760 -----------AAGUAAUUUUUUCAAGUGUCCAAUAUCAUCACAUAAACACGUUAUCGAGGGGGCUGGAGGCGACUCGGUGAGUUUUAUGUGUG--AGUGCAAGCAGCUU -----------...............((...((...(((.((((((((.((.((((((((..(.(....).)..))))))))))))))))))))--).))...))..... ( -25.30, z-score = -1.21, R) >droSec1.super_0 3989754 108 + 21120651 UGUAGAGAU-AUAAUAUUUUUUUGCAGUGCCUAGGAUCGUUCUGUAAGCACUUUAUAGAGGGGCUA-GGGGCGCCACUAUGAUUUUUAUGUGCGCUAGUGCAGGCAGCUU ((((((((.-........))))))))((.(((((..((.(((((((((...))))))))).)))))-)).))(((..(((((...)))))((((....)))))))..... ( -29.60, z-score = 0.16, R) >droYak2.chr3R 8792528 109 - 28832112 UCUUCAGAUAACAAUC-UUUUUUGCAGUGCUCGGAAUAGGUCUGUAAGCACUUUAUAGAGGGGCUAGGGGGCGCCACUAUGAUUUUUAUGUGCGCGAGUGCAGGCAGCUU .....((((....)))-)...((((((((((((((.....))))..))))))((((((.((.(((....))).)).))))))......((..(....)..)).))))... ( -31.40, z-score = -0.35, R) >droEre2.scaffold_4770 891359 109 + 17746568 UGUACAGAUAACAAUCUUUUUUUGCAGUGCUCGGGAUCGGUC-GUAAGCACUUUAUAGAGGGGCUAGGGGGCGCCACUGUGAUUUUUAUGUGCGCGAGUGCAGGCAGCUU .....((((....))))....((((((((((.(.((....))-.).))))))((((((.((.(((....))).)).))))))......((..(....)..)).))))... ( -30.80, z-score = 0.14, R) >droSim1.chr3R 10949713 108 + 27517382 UGCUGAGAU-ACAAUAUUUUUUU-CAGUGCCUAGGAUCGUUCUGUAAGCACUUUAUAGAGGGGCUAGGGGGCGCCACUAUGAUUUUUAUGUGCGCUAGUGCAGGCAGCUU .(((((((.-..........)))-))))((((.(((....)))....((((.((((((.((.(((....))).)).)))))).......))))((....))))))..... ( -31.10, z-score = -0.15, R) >droVir3.scaffold_13047 3155387 91 - 19223366 --------------UAUUGGGGCGGGACAUGUAGAGGGCGCCAGAGUGCGCGCAGCUUCCGGCAAAUCGCUGGCUCCUGCUCUAUUUAUAUGUGCAAGUGUGCAU----- --------------...((((((((((........(.((((......)))).).....(((((.....))))).)))))))))).....(((..(....)..)))----- ( -33.90, z-score = -0.38, R) >consensus UGU_GAGAU_ACAAUAUUUUUUUGCAGUGCCUAGGAUCGGUCUGUAAGCACUUUAUAGAGGGGCUAGGGGGCGCCACUAUGAUUUUUAUGUGCGCUAGUGCAGGCAGCUU ...........................((((.....................((((((.((.(((....))).)).))))))......((..(....)..)))))).... (-16.43 = -14.86 + -1.58)

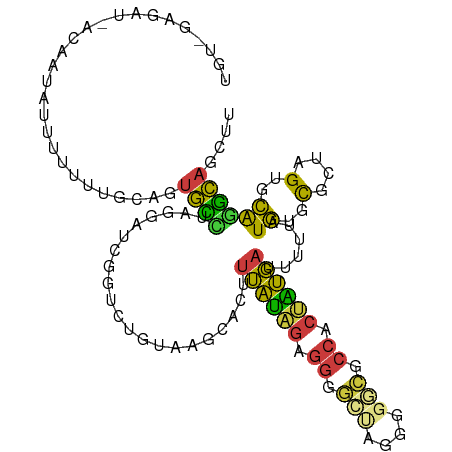

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:26 2011