| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,709,344 – 4,709,434 |
| Length | 90 |
| Max. P | 0.741670 |

| Location | 4,709,344 – 4,709,434 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.65 |
| Shannon entropy | 0.59820 |
| G+C content | 0.44461 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -8.91 |
| Energy contribution | -8.56 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
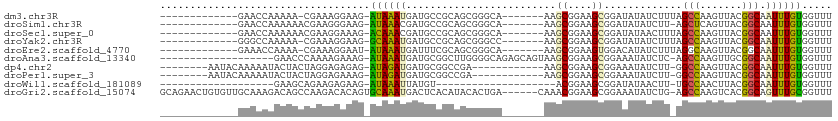
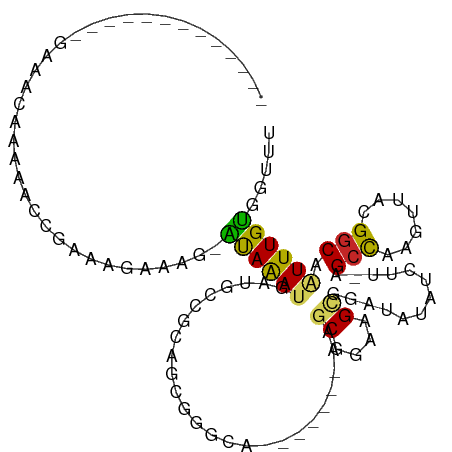
>dm3.chr3R 4709344 90 + 27905053 -------------GAACCAAAAA-CGAAAGGAAG-AUAAAUGAUGCCGCAGCGGGCA-------AAGCGGAAGCGGAUAUAUCUUUAGCCAAGUUACGGCAAUUUGUGGUUU -------------(((((.....-(....)....-((((((..(((((.(((.(((.-------..((....))(((....)))...)))..))).)))))))))))))))) ( -27.30, z-score = -3.01, R) >droSim1.chr3R 10927347 90 - 27517382 -------------GAACCAAAAAACGAAGGGAAG-AUAAACGAUGCCGCAGCGGGCA-------AAGCGGAAGCGGAUAUAUCUU-AGCUCAGUUACGGCAAUUUGUGGUUU -------------((((((.(((.(....)....-........(((((.(((((((.-------..((....))(((....))).-.)))).))).))))).))).)))))) ( -26.40, z-score = -2.51, R) >droSec1.super_0 3967772 91 - 21120651 -------------GAACCAAAAAACGAAGGAAAG-ACAAACGAUGCCGCAGCGGGCA-------AAGCGGAAGCGGAUAUAACUUUAGCCAAGUUACGGCAAUUUGUGGUUU -------------(((((......(....)....-(((((...(((((.(((.(((.-------..((....))(.......)....)))..))).))))).)))))))))) ( -25.40, z-score = -2.37, R) >droYak2.chr3R 8769156 90 + 28832112 -------------GGGCCAAAAA-CGAAAGGAAG-GCAAAUGAUGCCGCAGCGGGCC-------AAGCGGAAGCGGAUAUAUCUUUAGCCAAGUUACGGCAAUUUGUGGUUU -------------.((((.....-(....)....-((((((..(((((.(((.(((.-------..((....))(((....)))...)))..))).))))))))))))))). ( -27.70, z-score = -1.25, R) >droEre2.scaffold_4770 868302 90 - 17746568 -------------GAAACCAAAA-CGAAAGGAAU-AUAAAUGAUUUCGCAGCGGGCA-------AAGCGGAAGUGGACAUAUCUUUAGGCAAGUUACGGCAAUUUGUGGUUU -------------.(((((....-(....)....-((((((....(((.(((..((.-------..((....))(((....)))....))..))).)))..))))))))))) ( -16.60, z-score = -0.22, R) >droAna3.scaffold_13340 3478269 91 + 23697760 -------------------GAACCCAAAAGAAAG-AUAAAUGAUGCGGCUUGGGGCAGAGCAGUAAGCGGAAGCGGAAAUAUCUC-AGCCAAGUUGCGGCAAUUUGUGGUUU -------------------(((((..........-((((((..((((((((((.(.(((.......((....)).......))))-..))))))))))...))))))))))) ( -27.14, z-score = -1.83, R) >dp4.chr2 7738651 90 - 30794189 --------AAUACAAAAAUACUACUAGGAGAGAG-AUAGAUGAUGCGGCCGA------------AAGCGGAAGCGGAAAUAUCUU-GGCCAAGUUACGGCAAUUUGUGGUUU --------..((((((......(((.((...(((-(((.....(((..(((.------------...)))..)))....))))))-..)).)))........)))))).... ( -21.64, z-score = -1.88, R) >droPer1.super_3 1547397 90 - 7375914 --------AAUACAAAAAUACUACUAGGAGAAAG-AUAGAUGAUGCGGCCGA------------AAGCGGAAGCGGAAAUAUCUU-GGCCAAGUUACGGCAAUUUGUGGUUU --------..((((((......(((.((...(((-(((.....(((..(((.------------...)))..)))....))))))-..)).)))........)))))).... ( -21.24, z-score = -1.80, R) >droWil1.scaffold_181089 6368544 71 + 12369635 -------------------GAAGCAGAAGAGAAG-AUAAAUUAUGU--------------------ACGGAAGCGGAUAUAACUU-UGCCAACUUACGGCAAUUUGUGGUUU -------------------...(((((.......-.....((((((--------------------.((....)).))))))..(-((((.......))))))))))..... ( -13.20, z-score = -1.28, R) >droGri2.scaffold_15074 7498963 105 + 7742996 GCAGAACUGUGUUGCAAAGACAGCCAAGACACAGUGCAAAUGACUCACAUACACUGA------CAAACGGAAGCGGAAAUAUCUG-AGCCAAGUCACGGCAGUUUGCGGUUU (((((.((((((((......))))...(((.(((((...(((.....))).))))).------.....((...((((....))))-..))..)))...)))))))))..... ( -26.90, z-score = -0.49, R) >consensus _____________GAAACAAAAACCGAAAGAAAG_AUAAAUGAUGCCGCAGCGGGCA_______AAGCGGAAGCGGAUAUAUCUU_AGCCAAGUUACGGCAAUUUGUGGUUU ...................................((((((.........................((....)).............(((.......))).))))))..... ( -8.91 = -8.56 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:25 2011