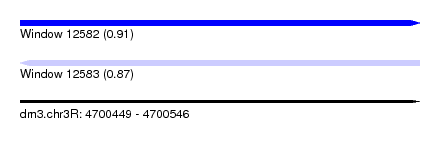
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,700,449 – 4,700,546 |
| Length | 97 |
| Max. P | 0.907492 |

| Location | 4,700,449 – 4,700,546 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Shannon entropy | 0.15909 |
| G+C content | 0.27725 |
| Mean single sequence MFE | -17.38 |
| Consensus MFE | -14.76 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
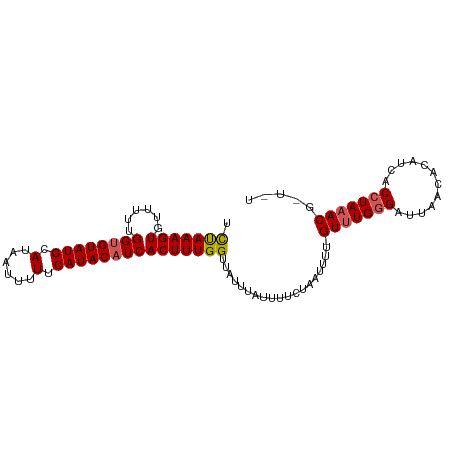
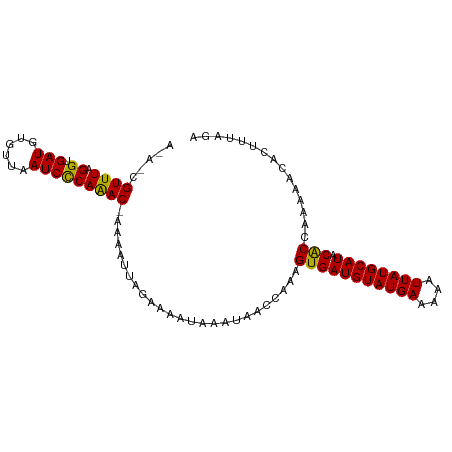
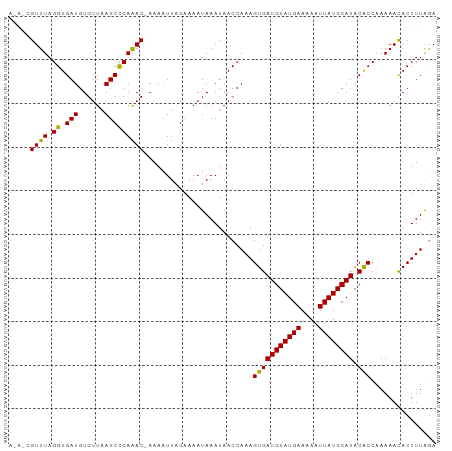
>dm3.chr3R 4700449 97 + 27905053 UUUAAAGUGUUUUUGGUGUAUGCAUAAAUUUUCAUACAUCACUUUGGUUAUUUAUUUUCAAAUUUU-GUUUGGGAUUAACACAUCACCUAAACGCUUU ...(((((((((.(((((((((.(......).)))))))))....(((.(((((.(..(((((...-)))))..).)))...)).))).))))))))) ( -20.80, z-score = -3.11, R) >droSim1.chr3R 10918730 93 - 27517382 UCUAAAGUGUUUUUGGCGUAUGCAUAAUUUUUCAUACAUCACUUUGGUUAUUUAUUUUCUAAUUUUUGUUUGGGAUUAACACAUCACCUAAAC----- .((((((((........(((((.(......).)))))..))))))))....................(((((((............)))))))----- ( -13.30, z-score = -1.01, R) >droSec1.super_0 3959086 93 - 21120651 UCUAAAGUGUUUUUGGUGUAUGCAUAAUUUUUCAUACAUCACUUUGGUUAUUUAUUUUCUAAAUUUUGUUUGGGAUUAACACAUCACCUAAAC----- .((((((((......(((((((.(......).)))))))))))))))....................(((((((............)))))))----- ( -16.70, z-score = -1.89, R) >droYak2.chr3R 8760242 97 + 28832112 UCCAAAGUGUUUUUGGUGUAUGCAUAAUUUUUCAUACAUCACUUUGGUUAUUUAUUUUCUAAUUUU-GUUUGGGAUUAACACAUCACCUAAACGCUCU .((((((((......(((((((.(......).)))))))))))))))...................-(((((((............)))))))..... ( -20.70, z-score = -3.12, R) >droEre2.scaffold_4770 859540 96 - 17746568 UCUAAAGUAUUUU-GGUGUAUGCAUAAUUUUUCAUACAUCACUUUGGUUAUUUAUUUGCUAAUUUU-GUCUGAGAUAAACACAUCACCUAAACGUUUU ...((((..(((.-((((.(((.......(((((.(((.....(((((.........)))))...)-)).)))))......))))))).)))..)))) ( -15.42, z-score = -1.28, R) >consensus UCUAAAGUGUUUUUGGUGUAUGCAUAAUUUUUCAUACAUCACUUUGGUUAUUUAUUUUCUAAUUUU_GUUUGGGAUUAACACAUCACCUAAACG_U_U .(((((((......((((((((.(......).)))))))))))))))....................(((((((............)))))))..... (-14.76 = -15.04 + 0.28)

| Location | 4,700,449 – 4,700,546 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Shannon entropy | 0.15909 |
| G+C content | 0.27725 |
| Mean single sequence MFE | -16.68 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4700449 97 - 27905053 AAAGCGUUUAGGUGAUGUGUUAAUCCCAAAC-AAAAUUUGAAAAUAAAUAACCAAAGUGAUGUAUGAAAAUUUAUGCAUACACCAAAAACACUUUAAA ((((.((((.((.(((......)))))....-........................(((((((((((....)))))))).)))...)))).))))... ( -17.30, z-score = -2.23, R) >droSim1.chr3R 10918730 93 + 27517382 -----GUUUAGGUGAUGUGUUAAUCCCAAACAAAAAUUAGAAAAUAAAUAACCAAAGUGAUGUAUGAAAAAUUAUGCAUACGCCAAAAACACUUUAGA -----((((.((.(((......))))))))).........................(((((((((((....)))))))).)))............... ( -14.60, z-score = -1.84, R) >droSec1.super_0 3959086 93 + 21120651 -----GUUUAGGUGAUGUGUUAAUCCCAAACAAAAUUUAGAAAAUAAAUAACCAAAGUGAUGUAUGAAAAAUUAUGCAUACACCAAAAACACUUUAGA -----((((.((.(((......)))))))))...(((((.....))))).......(((((((((((....)))))))).)))............... ( -15.20, z-score = -1.84, R) >droYak2.chr3R 8760242 97 - 28832112 AGAGCGUUUAGGUGAUGUGUUAAUCCCAAAC-AAAAUUAGAAAAUAAAUAACCAAAGUGAUGUAUGAAAAAUUAUGCAUACACCAAAAACACUUUGGA .....((((.((.(((......)))))))))-...................((((((((((((((((....))))))))..........)))))))). ( -21.20, z-score = -3.31, R) >droEre2.scaffold_4770 859540 96 + 17746568 AAAACGUUUAGGUGAUGUGUUUAUCUCAGAC-AAAAUUAGCAAAUAAAUAACCAAAGUGAUGUAUGAAAAAUUAUGCAUACACC-AAAAUACUUUAGA .....((((((((((.....)))))).))))-........................(((((((((((....)))))))).))).-............. ( -15.10, z-score = -1.10, R) >consensus A_A_CGUUUAGGUGAUGUGUUAAUCCCAAAC_AAAAUUAGAAAAUAAAUAACCAAAGUGAUGUAUGAAAAAUUAUGCAUACACCAAAAACACUUUAGA .....((((.((.(((......))))).............................(((((((((((....)))))))).)))...))))........ (-13.78 = -13.30 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:24 2011