
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,688,649 – 4,688,791 |
| Length | 142 |
| Max. P | 0.994138 |

| Location | 4,688,649 – 4,688,791 |
|---|---|
| Length | 142 |
| Sequences | 4 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.34406 |
| G+C content | 0.41296 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
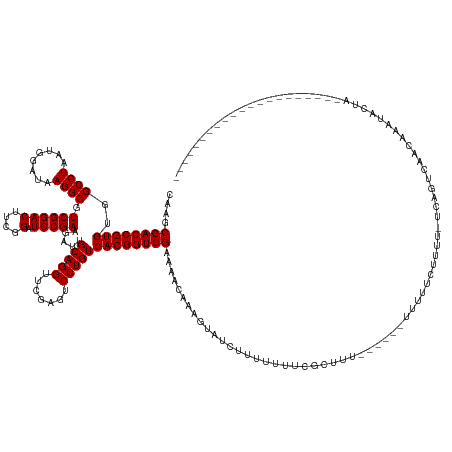
>dm3.chr3R 4688649 142 + 27905053 CAAGCGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAAACCAAAGUAUUAUUUUUUUUUUGUGAGAAACUUAUGUUUUGUUCUUUAAAAAAUUCAAUUUG----------------- ....((((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))))....((((.....(((((((....(...(((((....)))))...)...)))))))....))))----------------- ( -39.50, z-score = -1.89, R) >droPer1.super_3 1520029 131 - 7375914 CAAGCGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAAUAGAAAGUAUCCUUUUUUCGCUUU------UUUUUCUUUU-UCAGUCAACAAAUAUUA--------------------- .(((((((((((..((((........)))).((((((....).))))).....(((((.......))))))))))))(((..(((((....)))))))))))).------..........-.................--------------------- ( -37.50, z-score = -1.66, R) >dp4.chr2 7709194 131 - 30794189 CAAGCGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAACACAAAGUAUCUUUUUUUCGCUUU------UUUUUCUUUU-UCAGUCAACCAAUAUUA--------------------- .(((((((((((..((((........)))).((((((....).))))).....(((((.......))))))))))))(((...((((...))))..))))))).------..........-.................--------------------- ( -35.20, z-score = -1.16, R) >droEre2.scaffold_4770 851687 153 - 17746568 CAAGCGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGACACUAAAGUAAUUUUUUUUUUUUUU------UUACUUUUUUGUAAGAAAAUUCUUACUUUCUCAAAAAAAACUAAAUAUG ...(((((((((..((((........)))).((((((....).))))).....(((((.......))))))))))))).)...(((((((.........(((((------((((......)))))))))...))))))).................... ( -40.90, z-score = -2.48, R) >consensus CAAGCGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAAAACAAAGUAUCUUUUUUUCGCUUU______UUUUUCUUUU_UCAGUCAACAAAUACUA_____________________ ....((((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))))................................................................................. (-33.00 = -33.00 + 0.00)

| Location | 4,688,649 – 4,688,791 |
|---|---|
| Length | 142 |
| Sequences | 4 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.34406 |
| G+C content | 0.41296 |
| Mean single sequence MFE | -35.81 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
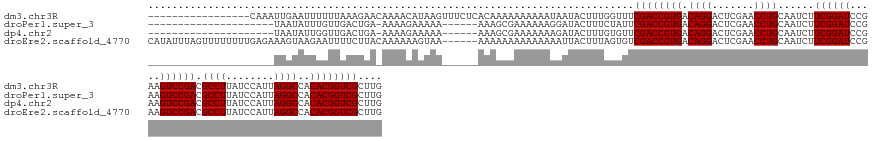
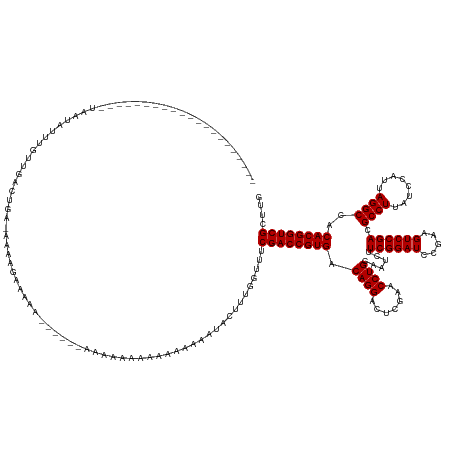
>dm3.chr3R 4688649 142 - 27905053 -----------------CAAAUUGAAUUUUUUAAAGAACAAAACAUAAGUUUCUCACAAAAAAAAAAUAAUACUUUGGUUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCGCUUG -----------------((((....(((((((........((((....)))).........))))))).....))))....((((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))).... ( -30.93, z-score = -1.66, R) >droPer1.super_3 1520029 131 + 7375914 ---------------------UAAUAUUUGUUGACUGA-AAAAGAAAAA------AAAGCGAAAAAAGGAUACUUUCUAUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCGCUUG ---------------------.................-..........------.(((((((.((((....))))...)))(((((((.((((.......))))......((((((.....)))))).((((........))))..))))))))))). ( -34.80, z-score = -2.39, R) >dp4.chr2 7709194 131 + 30794189 ---------------------UAAUAUUGGUUGACUGA-AAAAGAAAAA------AAAGCGAAAAAAAGAUACUUUGUGUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCGCUUG ---------------------.....(..(....)..)-..........------.(((((((..((((...))))...)))(((((((.((((.......))))......((((((.....)))))).((((........))))..))))))))))). ( -35.30, z-score = -2.14, R) >droEre2.scaffold_4770 851687 153 + 17746568 CAUAUUUAGUUUUUUUUGAGAAAGUAAGAAUUUUCUUACAAAAAAGUAA------AAAAAAAAAAAAAAUUACUUUAGUGUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCGCUUG ....((((.((((((.(((((((((....))))))))).)))))).)))------).......................(.((((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))))... ( -42.20, z-score = -4.01, R) >consensus _____________________UAAUAUUUGUUGACUGA_AAAAGAAAAA______AAAAAAAAAAAAAAAUACUUUGGUUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCGCUUG .................................................................................((((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))).... (-29.50 = -29.50 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:22 2011