
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,687,638 – 4,687,697 |
| Length | 59 |
| Max. P | 0.898793 |

| Location | 4,687,638 – 4,687,697 |
|---|---|
| Length | 59 |
| Sequences | 9 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 67.59 |
| Shannon entropy | 0.63861 |
| G+C content | 0.42533 |
| Mean single sequence MFE | -11.57 |
| Consensus MFE | -8.72 |
| Energy contribution | -7.76 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.41 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
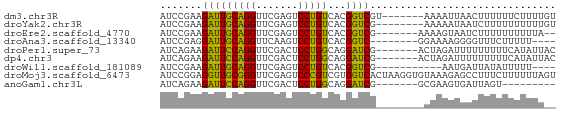
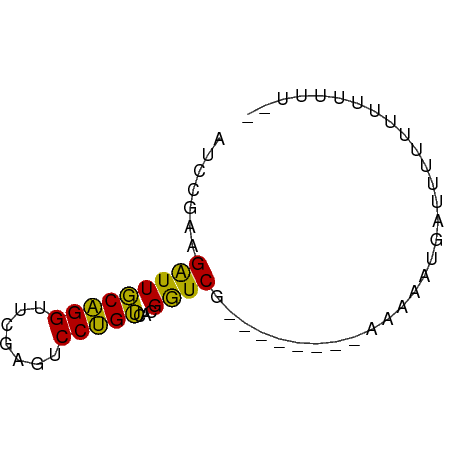
>dm3.chr3R 4687638 59 + 27905053 AUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGU-------AAAAUUAACUUUUUUCUUUUGU ...(((.....(((((.......))))).....))).-------...................... ( -8.50, z-score = 0.28, R) >droYak2.chr3R 8751165 58 + 28832112 AUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG--------AAAAAUAAUCUUUUUUUUUUGU ....((((((((....(((((..((......)))))--------))...))))))))......... ( -10.70, z-score = -0.46, R) >droEre2.scaffold_4770 850705 57 - 17746568 AUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG-------AAAAGUAAUCUUUUUUUUUUA-- ....(((((((((...(((((..((......)))))-------))..)))))))))........-- ( -14.30, z-score = -2.13, R) >droAna3.scaffold_13340 3460195 54 + 23697760 AUCCGAGGAUUGCAGGUUCAAGUCCUGUCACGGUC--------GGAAAAGGGGUUUCUUUUU---- .(((((.....(((((.......))))).....))--------)))((((((....))))))---- ( -13.20, z-score = -0.32, R) >droPer1.super_73 71716 59 + 267513 AUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-------ACUAGAUUUUUUUUUCAUAUUAC ....(((((..(((((.......))))).((((((.-------....)))))).)))))....... ( -12.10, z-score = -1.23, R) >dp4.chr3 1794702 59 - 19779522 AUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-------ACUAGAUUUUUUUUUCAUAUUAC ....(((((..(((((.......))))).((((((.-------....)))))).)))))....... ( -12.10, z-score = -1.23, R) >droWil1.scaffold_181089 5049323 51 + 12369635 AUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG-----------AAUGAUUAUAUUUUU---- ...(((.....(((((.......))))).....)))-----------...............---- ( -9.00, z-score = 0.19, R) >droMoj3.scaffold_6473 8003058 66 - 16943266 AUCCGGAGGUUGCGGGUUCGAGUCCCGUCGUGGUCACUAAGGUGUAAAGAGCCUUUCUUUUUUAGU ....((((((((((((.......)))))......(((....))).....))))))).......... ( -14.30, z-score = 0.50, R) >anoGam1.chr3L 7649581 50 - 41284009 AUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-------GCGAAGUGAUUAGU--------- ....(..(((((((((.......)))))...)))).-------.)............--------- ( -9.90, z-score = 0.70, R) >consensus AUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCG________AAAAAUGAUUUUUUUUUUUU__ .......(((((((((.......)))))...))))............................... ( -8.72 = -7.76 + -0.96)
| Location | 4,687,638 – 4,687,697 |
|---|---|
| Length | 59 |
| Sequences | 9 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 67.59 |
| Shannon entropy | 0.63861 |
| G+C content | 0.42533 |
| Mean single sequence MFE | -9.84 |
| Consensus MFE | -4.72 |
| Energy contribution | -4.63 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
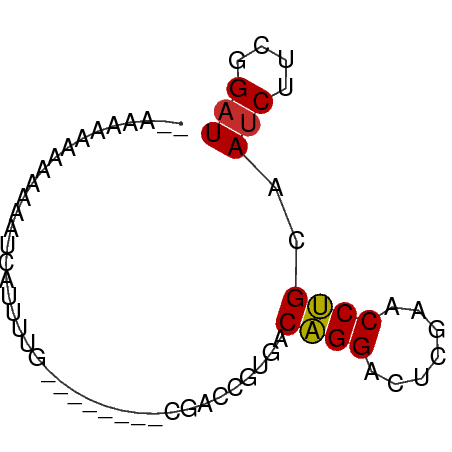
>dm3.chr3R 4687638 59 - 27905053 ACAAAAGAAAAAAGUUAAUUUU-------ACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAU ......................-------.(((...((.((((.......))))))....)))... ( -8.00, z-score = -0.03, R) >droYak2.chr3R 8751165 58 - 28832112 ACAAAAAAAAAAGAUUAUUUUU--------CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAU ....................((--------(((...((.((((.......))))))....))))). ( -9.30, z-score = -0.64, R) >droEre2.scaffold_4770 850705 57 + 17746568 --UAAAAAAAAAAGAUUACUUUU-------CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAU --...................((-------(((...((.((((.......))))))....))))). ( -9.30, z-score = -0.92, R) >droAna3.scaffold_13340 3460195 54 - 23697760 ----AAAAAGAAACCCCUUUUCC--------GACCGUGACAGGACUUGAACCUGCAAUCCUCGGAU ----................(((--------((...((.((((.......))))))....))))). ( -12.20, z-score = -2.07, R) >droPer1.super_73 71716 59 - 267513 GUAAUAUGAAAAAAAAAUCUAGU-------CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAU .................((((((-------((((((.....))).))))..))))).......... ( -11.20, z-score = -1.44, R) >dp4.chr3 1794702 59 + 19779522 GUAAUAUGAAAAAAAAAUCUAGU-------CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAU .................((((((-------((((((.....))).))))..))))).......... ( -11.20, z-score = -1.44, R) >droWil1.scaffold_181089 5049323 51 - 12369635 ----AAAAAUAUAAUCAUU-----------CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAU ----............(((-----------(((...((.((((.......))))))....)))))) ( -9.90, z-score = -1.33, R) >droMoj3.scaffold_6473 8003058 66 + 16943266 ACUAAAAAAGAAAGGCUCUUUACACCUUAGUGACCACGACGGGACUCGAACCCGCAACCUCCGGAU ......(((((.....)))))...((...((....))(.((((.......))))).......)).. ( -9.00, z-score = 0.96, R) >anoGam1.chr3L 7649581 50 + 41284009 ---------ACUAAUCACUUCGC-------CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAU ---------..............-------.((.....(((((.......))))).....)).... ( -8.50, z-score = 0.16, R) >consensus __AAAAAAAAAAAAUCAUUUUG________CGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAU .......................................((((.......))))..(((....))) ( -4.72 = -4.63 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:20 2011