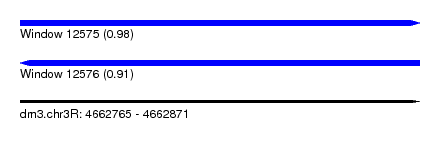
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,662,765 – 4,662,871 |
| Length | 106 |
| Max. P | 0.982153 |

| Location | 4,662,765 – 4,662,871 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.33 |
| Shannon entropy | 0.38901 |
| G+C content | 0.50887 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -23.80 |
| Energy contribution | -26.90 |
| Covariance contribution | 3.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4662765 106 + 27905053 UGGCCAGAUCCGGCUGAUGCUGCUGCUGCUGCUGGGGCAGAAGAUUCAGACUGGAAUCGAAGAGAUUCUUCUGCAGCAGCAGCUCGUGGUAGUAGUAGGGAGUGGU ..((((..(((..(((.((((((..(.(((((((..((((((((.((...((........)).)).))))))))..)))))))..)..)))))).)))))).)))) ( -48.60, z-score = -3.20, R) >droSim1.chr3R 10882195 100 - 27517382 UGGCCAGAUCCGG------CUGCUGCUGCUGCUGGGGCAGAAGAUUGAGACUGGAAUCGAAGAGAUUCUUCUGCAGCAGGAGCUCGUGGUAGUAGUAGGGCGUGGU ..((((..(((.(------(((((((((((.(((..((((((((((((........))))......))))))))..))).)))....))))))))).)))..)))) ( -42.70, z-score = -2.46, R) >droWil1.scaffold_181089 6324798 95 + 12369635 -------UGCUGCUUGAGAUUAUUGUU-CUUCUCUUGCAGUAGAUUCAAACUUGAAUCGAAAACGUUCUUCUGGAGCAG---UUCAUGAUAGUAAUAGGGCGUGGU -------((((((..((((........-..))))..))))))((((((....))))))....(((((((.(((...((.---....)).)))....)))))))... ( -27.40, z-score = -1.95, R) >consensus UGGCCAGAUCCGG_UGA__CUGCUGCUGCUGCUGGGGCAGAAGAUUCAGACUGGAAUCGAAGAGAUUCUUCUGCAGCAG_AGCUCGUGGUAGUAGUAGGGCGUGGU ..((((..(((.((((..(((((.((((((((((...((((((((((......)))))(((....))))))))))))))))))..)))))..)))).)))..)))) (-23.80 = -26.90 + 3.10)


| Location | 4,662,765 – 4,662,871 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.33 |
| Shannon entropy | 0.38901 |
| G+C content | 0.50887 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -17.11 |
| Energy contribution | -18.53 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4662765 106 - 27905053 ACCACUCCCUACUACUACCACGAGCUGCUGCUGCAGAAGAAUCUCUUCGAUUCCAGUCUGAAUCUUCUGCCCCAGCAGCAGCAGCAGCAUCAGCCGGAUCUGGCCA .....................(((((((((((((.((((.....))))(((((......)))))...(((....)))))))))))))).)).((((....)))).. ( -39.20, z-score = -3.82, R) >droSim1.chr3R 10882195 100 + 27517382 ACCACGCCCUACUACUACCACGAGCUCCUGCUGCAGAAGAAUCUCUUCGAUUCCAGUCUCAAUCUUCUGCCCCAGCAGCAGCAGCAG------CCGGAUCUGGCCA .....(((............((.(((.((((((((((.(((((.....)))))...)))........(((....)))))))))).))------))).....))).. ( -27.63, z-score = -1.58, R) >droWil1.scaffold_181089 6324798 95 - 12369635 ACCACGCCCUAUUACUAUCAUGAA---CUGCUCCAGAAGAACGUUUUCGAUUCAAGUUUGAAUCUACUGCAAGAGAAG-AACAAUAAUCUCAAGCAGCA------- ........................---(((((..(((.(((((....)).)))..((((...(((......)))...)-))).....)))..)))))..------- ( -15.40, z-score = -0.50, R) >consensus ACCACGCCCUACUACUACCACGAGCU_CUGCUGCAGAAGAAUCUCUUCGAUUCCAGUCUGAAUCUUCUGCCCCAGCAGCAGCAGCAG__UCA_CCGGAUCUGGCCA .......................((((((((((..((((.....))))(((((......)))))........))))))))))........................ (-17.11 = -18.53 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:18 2011