| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,615,476 – 4,615,657 |
| Length | 181 |
| Max. P | 0.981742 |

| Location | 4,615,476 – 4,615,657 |
|---|---|
| Length | 181 |
| Sequences | 6 |
| Columns | 183 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Shannon entropy | 0.18186 |
| G+C content | 0.42204 |
| Mean single sequence MFE | -56.98 |
| Consensus MFE | -43.96 |
| Energy contribution | -47.05 |
| Covariance contribution | 3.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
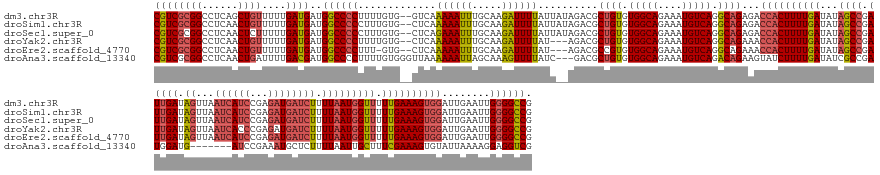
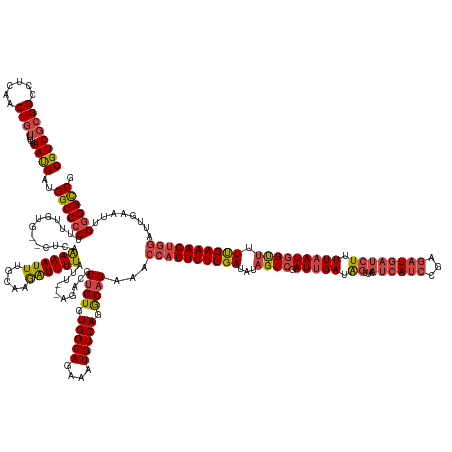
>dm3.chr3R 4615476 181 + 27905053 CGUCGCGGCCUCAGCUGUUUUUGAUGAUGGCCCCUUUUGUG--GUCAAAAAUUUGCAAGAUUUUAUUAUAGACGCUGUGUGGCAGAAAUGUCAGGCAGAGACCACUUUUGAUAUAGCCGAUUGAUAGUUAAUCAUCCGAGAUGAUCUUUUAAUGGUUUUUGAAAGUGGAUUGAAUUGGGGCCG (((((((((....)))))....))))..(((((((((((..--(........)..)))))............(.((((.(((((....))))).)))).).(((((((..(...((((.(((((.((...((((((...)))))))).))))))))).)..)))))))........)))))). ( -63.10, z-score = -4.14, R) >droSim1.chr3R 10840827 181 - 27517382 CGUCGCGGCCUCAACUGUUUUUGAUGAUGGCCCCCUUUGUG--CUCAAAAAUUUGCAAGAUUUUAUUAUAGACGCUGUGUGGCAGAAAUGUCAGGCAGAGACCACUUUUGAUAUAGCCGAUUGAUAGUUAAUCAUCCGAGAUGAUCUUUUAAUGGUUUUUGAAAGUGGAUUGAAUUGGGGCCG .....(((((((((((((((.((((((..((....((((..--..)))).....))......)))))).)))).((((.(((((....))))).)))))).(((((((..(...((((.(((((.((...((((((...)))))))).))))))))).)..)))))))......))))))))) ( -57.70, z-score = -2.99, R) >droSec1.super_0 3878739 181 - 21120651 CGUCGCGGCCUCAACUCUUUUUGAUGAUGGCCCCCUUUGUG--CUCAGAAAUUUGCAAGAUUUUAUUAUAGACGCUGUGUGGCAGAAAUGUCAGGCAGAGACCACUUUUGAUAUAGCCGAUUGAUAGUUAAUCAUCCGAGAUGAUCUUUUAAUGGUUUUUGAAAGUGGAUUGAAUUGGGGCCG .....(((((((((.(((((((((..(..(......)..).--.))))))).....................(.((((.(((((....))))).)))).).(((((((..(...((((.(((((.((...((((((...)))))))).))))))))).)..)))))))...)).))))))))) ( -56.50, z-score = -2.48, R) >droYak2.chr3R 8677030 178 + 28832112 CGUCGCGGCCUCAACUGUUUUUGAUGAUGGCCCCUUUUGUG--CUCAAAAAUUUGCAAGAUUUUAU---AGACGCUGUGUGGCAGAAAUGUCAGGCAGAAACCACUUUUGAUAUAGCCGAUUGAUAGUUAAUCACCCGAGAUGAUCUUUUAAUGGUUUUUGAAAGUGGAUUGAAUUGGGGCCG ((((((((......))))....))))..((((((.....((--(..........))).(((((.((---.....((((.(((((....))))).))))...(((((((..(...((((.(((((.((...(((......)))...)).))))))))).)..))))))))).))))))))))). ( -56.50, z-score = -3.01, R) >droEre2.scaffold_4770 775439 177 - 17746568 CGUCGCGGCCUCAACUGUUUUUGAUGAUGGCCCCUUU-GUG--CUCAAAAAUUUGCAAGAUUUUAU---AGACGCCGUGUGGCAGAAAUGUCAGGCAGAAACCACUUUUGAUAUAGCCGAUUGAUAGUUAAUCAUCCGAGAUGAUCUUUUAAUGGUUUUUGAAAGUGGAUUGAAUUGGGGCCG ((((((((......))))....))))..((((((...-(((--((..((((((.....))))))..---)).))).((.(((((....))))).)).....(((((((..(...((((.(((((.((...((((((...)))))))).))))))))).)..)))))))........)))))). ( -55.00, z-score = -2.54, R) >droAna3.scaffold_13340 11025155 173 + 23697760 CGUCGCGGCCUCAACUGAUUUUGACGAUGGCCCCUUUUGUGGGUUAAAAAAUUAGCAAAGUUUUAUC---GACGCUGUGUGGCAGAAAUGUCAGACAGAAGUAUCUUUUGAUAUCGCCGAUGGAUG-------AUCCGAAAUGCUCUUUUAAUUGCUUUCGAAAGUGUAUUAAAAGGAGGUCG .....(((((((..((((((((.....(((((((....).)))))).))))))))......((((((---(.((((((.(((((....))))).))))..(((((....))))))).)))))))..-------............((((((((.((...(....).))))))))))))))))) ( -53.10, z-score = -2.48, R) >consensus CGUCGCGGCCUCAACUGUUUUUGAUGAUGGCCCCUUUUGUG__CUCAAAAAUUUGCAAGAUUUUAUU__AGACGCUGUGUGGCAGAAAUGUCAGGCAGAAACCACUUUUGAUAUAGCCGAUUGAUAGUUAAUCAUCCGAGAUGAUCUUUUAAUGGUUUUUGAAAGUGGAUUGAAUUGGGGCCG ((((((((......))))....))))..((((((.............((((((.....))))))..........((((.(((((....))))).))))...((((((((((...((((.(((((.((...(((......)))...)).))))))))).))))))))))........)))))). (-43.96 = -47.05 + 3.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:15 2011