
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,773,773 – 6,773,831 |
| Length | 58 |
| Max. P | 0.631252 |

| Location | 6,773,773 – 6,773,831 |
|---|---|
| Length | 58 |
| Sequences | 9 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.57531 |
| G+C content | 0.49304 |
| Mean single sequence MFE | -12.02 |
| Consensus MFE | -8.26 |
| Energy contribution | -8.09 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.35 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6773773 58 + 23011544 GAUCAGCUGACUG--UCAGCUGUAUGUAAACAAGUUAAAAUGCCGG-------GUUCCUCUCCUUCG ...((((((....--.))))))......................((-------(......))).... ( -10.20, z-score = -0.51, R) >droSim1.chr2L 6574238 58 + 22036055 GAUCAGCUGACUG--UCAGCUGUAUGUAAACAAGUUAAAAUGCCGG-------GUUCCUCUCCUUCG ...((((((....--.))))))......................((-------(......))).... ( -10.20, z-score = -0.51, R) >droSec1.super_3 2312410 58 + 7220098 GAUCAGCUGACUG--UCAGCUGUAUGUAAACAAGUUAAAAUGCCGG-------GUUCCUCUCCUUCG ...((((((....--.))))))......................((-------(......))).... ( -10.20, z-score = -0.51, R) >droYak2.chr2L 16188772 57 - 22324452 GAUCAGCUGACUG--CCAGCUGUAUGUAAACAAGUUAAAACGCCGG-------GUUC-UCUCCUUCG ...((((((....--.))))))......................((-------(...-...)))... ( -9.80, z-score = -0.03, R) >droEre2.scaffold_4929 15689910 57 + 26641161 GAUCAGCUGACUG--CCAGCUGUAUGUAAACAAGUUAAAAUGCCGG-------GUUC-UCUUCCUUG ...((((((....--.))))))......................((-------(...-...)))... ( -10.40, z-score = -0.24, R) >droAna3.scaffold_12916 8351518 61 + 16180835 --UCAGCUGU-UU-GCCAGCUGAGCGUAAACACU-CUGCAUGCCGGCAAUUGUGCUC-UCUCCGGCA --(((((((.-..-..)))))))(((........-.))).((((((...........-...)))))) ( -18.44, z-score = -0.44, R) >dp4.chr4_group1 3878892 55 + 5278887 ---CGGCUGCGUC-UACAGCUGAUUCUGCACCGG-CAAAAUGUAGAA------GCUC-UCUCCCGCG ---((((((....-..)))))).(((((((....-.....)))))))------((..-......)). ( -14.30, z-score = -0.49, R) >droPer1.super_5 5422116 55 - 6813705 ---CGGCUGCGUC-UACAGCUGAUUCUGCACCGG-CAAAAUGUAGAA------GCUC-UCUCCUGCG ---((((((....-..)))))).(((((((....-.....)))))))------((..-......)). ( -14.90, z-score = -0.76, R) >droWil1.scaffold_180772 7398838 64 + 8906247 GAUCAGCUGUUUGGUCCAGCUGUUUAUAAACAAUUCAAAAUGCCGGC---UAUGCUGUUUUUCUUUG ...((((((.......)))))).....................((((---...)))).......... ( -9.70, z-score = 0.31, R) >consensus GAUCAGCUGACUG__CCAGCUGUAUGUAAACAAGUUAAAAUGCCGG_______GUUC_UCUCCUUCG ...((((((.......))))))............................................. ( -8.26 = -8.09 + -0.17)

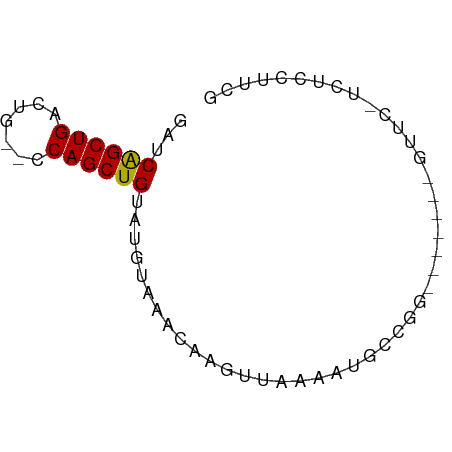
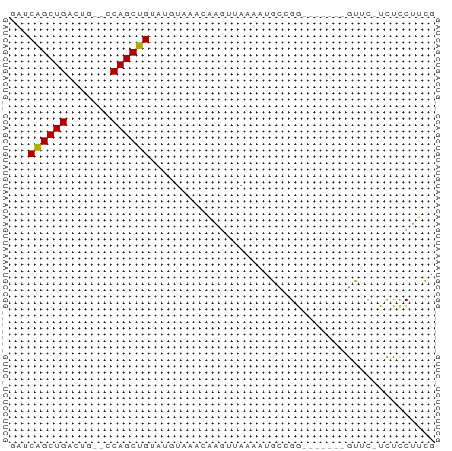
| Location | 6,773,773 – 6,773,831 |
|---|---|
| Length | 58 |
| Sequences | 9 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.57531 |
| G+C content | 0.49304 |
| Mean single sequence MFE | -13.63 |
| Consensus MFE | -8.23 |
| Energy contribution | -7.99 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6773773 58 - 23011544 CGAAGGAGAGGAAC-------CCGGCAUUUUAACUUGUUUACAUACAGCUGA--CAGUCAGCUGAUC .(((.(((..(((.-------.......)))..))).))).....((((((.--....))))))... ( -12.10, z-score = -0.88, R) >droSim1.chr2L 6574238 58 - 22036055 CGAAGGAGAGGAAC-------CCGGCAUUUUAACUUGUUUACAUACAGCUGA--CAGUCAGCUGAUC .(((.(((..(((.-------.......)))..))).))).....((((((.--....))))))... ( -12.10, z-score = -0.88, R) >droSec1.super_3 2312410 58 - 7220098 CGAAGGAGAGGAAC-------CCGGCAUUUUAACUUGUUUACAUACAGCUGA--CAGUCAGCUGAUC .(((.(((..(((.-------.......)))..))).))).....((((((.--....))))))... ( -12.10, z-score = -0.88, R) >droYak2.chr2L 16188772 57 + 22324452 CGAAGGAGA-GAAC-------CCGGCGUUUUAACUUGUUUACAUACAGCUGG--CAGUCAGCUGAUC .(((.((((-((((-------.....)))))..))).))).....((((((.--....))))))... ( -12.50, z-score = -0.38, R) >droEre2.scaffold_4929 15689910 57 - 26641161 CAAGGAAGA-GAAC-------CCGGCAUUUUAACUUGUUUACAUACAGCUGG--CAGUCAGCUGAUC ((((...((-((..-------......))))..))))........((((((.--....))))))... ( -10.70, z-score = -0.22, R) >droAna3.scaffold_12916 8351518 61 - 16180835 UGCCGGAGA-GAGCACAAUUGCCGGCAUGCAG-AGUGUUUACGCUCAGCUGGC-AA-ACAGCUGA-- ((((((..(-........)..))))))....(-((((....)))))(((((..-..-.)))))..-- ( -24.00, z-score = -2.10, R) >dp4.chr4_group1 3878892 55 - 5278887 CGCGGGAGA-GAGC------UUCUACAUUUUG-CCGGUGCAGAAUCAGCUGUA-GACGCAGCCG--- .(..((((.-...)------)))..)((((((-(....)))))))..(((((.-...)))))..--- ( -14.50, z-score = 0.25, R) >droPer1.super_5 5422116 55 + 6813705 CGCAGGAGA-GAGC------UUCUACAUUUUG-CCGGUGCAGAAUCAGCUGUA-GACGCAGCCG--- .(.(((((.-...)------)))).)((((((-(....)))))))..(((((.-...)))))..--- ( -13.90, z-score = 0.13, R) >droWil1.scaffold_180772 7398838 64 - 8906247 CAAAGAAAAACAGCAUA---GCCGGCAUUUUGAAUUGUUUAUAAACAGCUGGACCAAACAGCUGAUC .......((((((..((---(........)))..)))))).....((((((.......))))))... ( -10.80, z-score = -0.71, R) >consensus CGAAGGAGA_GAAC_______CCGGCAUUUUAACUUGUUUACAUACAGCUGG__CAGUCAGCUGAUC .......................((((........))))......(((((((.....)))))))... ( -8.23 = -7.99 + -0.24)
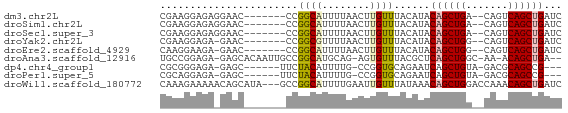
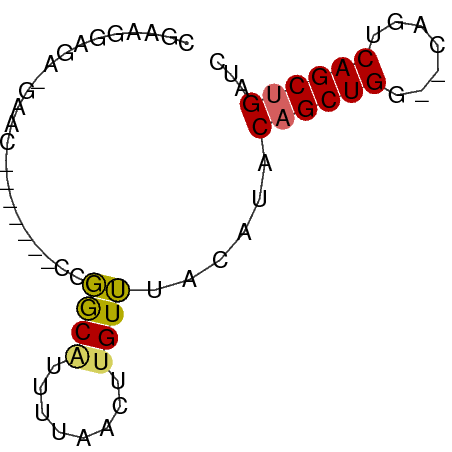
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:59 2011