
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,578,076 – 4,578,175 |
| Length | 99 |
| Max. P | 0.847531 |

| Location | 4,578,076 – 4,578,175 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 64.19 |
| Shannon entropy | 0.70323 |
| G+C content | 0.48100 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -9.49 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4578076 99 - 27905053 ----UUCUACAAUUUAUAAAAUUAG-GGGUCUUUUAGUGCCCAUGCUGCAUCA----UACUCACAGCAAUAGGGGCUGACCCAGCUCAUCGGCCAGUUCCCCGUAGAC ----.(((((((((.....)))).(-(((.((....(.(((..(((((.....----......)))))....((((((...))))))...))))))..))))))))). ( -27.00, z-score = -1.48, R) >droSim1.chr3R 10800665 99 + 27517382 ----UUCUACAAUUUAUAAAAUUAG-GGGACUUUCAGUGCCCAUGUUGCAUCA----UACUCACAGCAAUAGGGGCUGACCCAGCUCGUCGGCCAGUUCCCCGUAGAC ----.(((((((((.....)))).(-((((((........((.((((((....----........))))))))(((((((.......)))))))))))))).))))). ( -32.00, z-score = -3.04, R) >droSec1.super_0 3841862 92 + 21120651 -----------AUUUAUAAAAUUAG-GGGACUUUCAGUGCCCAUGUUGCAUCA----UACUCACAGCAAUAGGGGCUGACCCAGCUCAUCGGCCAGUUCCCCGUAGAC -----------.............(-((((((.(((((.(((.((((((....----........))))))))))))))....(((....))).)))))))....... ( -26.50, z-score = -1.56, R) >droYak2.chr3R 8639378 91 - 28832112 -----------GUAUA-AAGGGUAG-GGGACUUUUAGUCCAAAAAUUUCGUCG----UACUCACAGCAAUAGGGGCUGGCCGAGCUCAUCGGCCAGUUCCCCGUAGAC -----------.....-..(((((.-(((((.....))))...........).----))))).........((((((((((((.....))))))))..))))...... ( -32.50, z-score = -3.01, R) >droEre2.scaffold_4770 738319 103 + 17746568 AUGUUAAUACAGUUCAUAAAAUUAG-GGGACUUUUAGUCCUAGUAUUGCAUCG----UACUCACAGCAAUAGGGGCUGGCCCAGCUCAUCGGCCAGUUCCCCGUAGAC .......(((...............-(((((.....)))))..((((((....----........))))))((((((((((.........))))))..)))))))... ( -29.60, z-score = -1.89, R) >dp4.chr2 27227339 91 - 30794189 -----------AUUAAUAAAGUUA--CAACCGCUCUCCAUCAUCUGUGUC----AUCCACUCACAGCAGCAGGGGCUGCCCCAAUUCAUCGGCCAGCUCGCCGUAAAC -----------..........(((--(....(((.........(((((..----.......))))).)))..((((((.((.........)).))))))...)))).. ( -17.50, z-score = -0.66, R) >droPer1.super_6 2518186 91 - 6141320 -----------AUUAAUAAAGUUA--CAACCGCUCUCCAUCAUCUGUGUC----AUCCACUCACAGCAGCAGGGGCUGCCCCAAUUCAUCGGCCAGCUCGCCGUAAAC -----------..........(((--(....(((.........(((((..----.......))))).)))..((((((.((.........)).))))))...)))).. ( -17.50, z-score = -0.66, R) >droWil1.scaffold_181089 4577099 91 - 12369635 ----------GAAUCAUCAAAUUG--UACUCCGCCCCCGUCAUUUUAAUA----AU-UACUCACAACAAAAGCGGUUGUCCCAAUUCGUUAGCCAACUCGCCGUAGAC ----------..............--..((.((.....((.(((.....)----))-.))...........(((((((...............)))).))))).)).. ( -5.86, z-score = 1.51, R) >droMoj3.scaffold_6540 13154028 96 - 34148556 ----------AGAAUA-CAUGUUGAUUAUUUCGAUCUGAUUGCUAUAUCAUCG-AACUACUCACAGUAGCAGUGGCUGUCCCAACUCGUCGGCCAGCUCACCGUAAAC ----------....((-(.(((.((.((.(((((..((((......)))))))-)).)).))))))))((.(((((((.((.........)).)))).))).)).... ( -22.00, z-score = -1.05, R) >droGri2.scaffold_14906 10732795 96 + 14172833 -----------AAUUAUUAUAUUAUCUGCCCAGAUAUAAUC-UUGUGACAUCGUCAUUACUCACAGCAGCAGUGGCUGUCCCAGUUCAUCGGCCAGCUCGCCGUAGAC -----------.............(((((............-.(((((............)))))(((((..((((((...........))))))))).)).))))). ( -19.70, z-score = -0.45, R) >consensus ___________AUUUAUAAAAUUAG_GGGACUUUCAGUACCAAUGUUGCAUCG____UACUCACAGCAAUAGGGGCUGACCCAGCUCAUCGGCCAGCUCCCCGUAGAC ....................................................................((.(((((((.((.........)).)))).))).)).... ( -9.49 = -8.68 + -0.81)

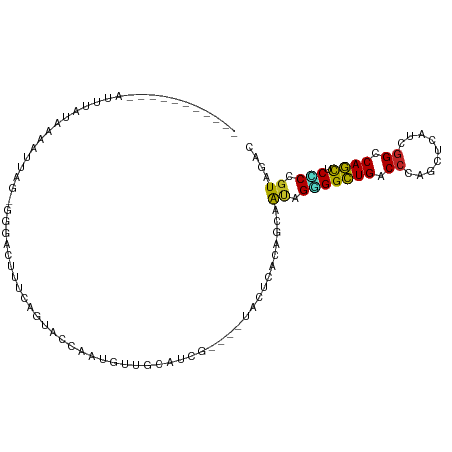
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:08 2011