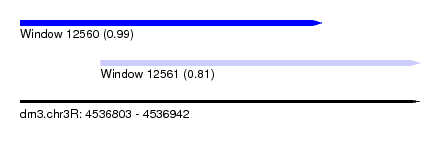
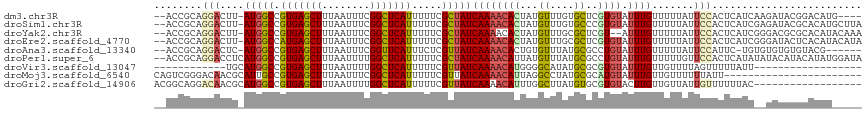
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,536,803 – 4,536,942 |
| Length | 139 |
| Max. P | 0.990856 |

| Location | 4,536,803 – 4,536,908 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Shannon entropy | 0.53652 |
| G+C content | 0.45143 |
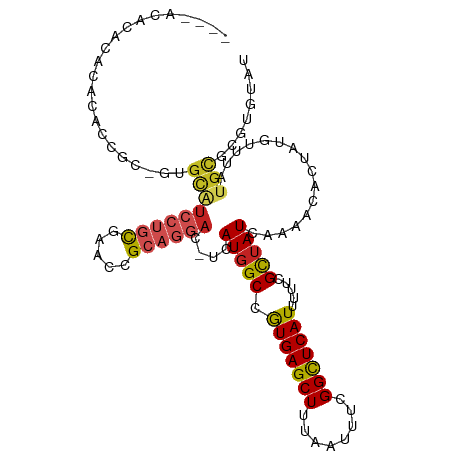
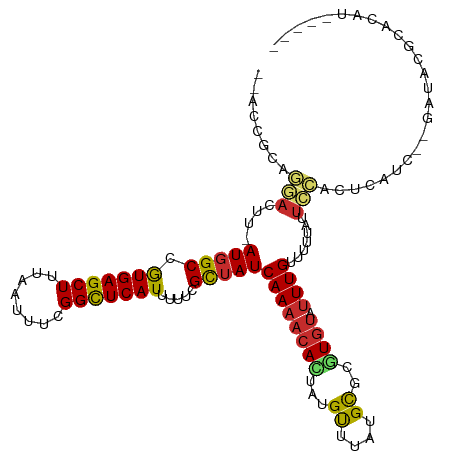
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.64 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
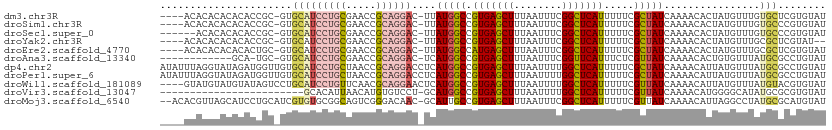
>dm3.chr3R 4536803 105 + 27905053 ----ACACACACACACCGC-GUGCAUCCUGCGAACCGCAGGAC-UUAUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGUGCUCGUGUAU ----((((.((((.((...-(((..(((((((...))))))).-..(((((.(((((((........))))))).....))))).....)))...)).))))...)))).. ( -30.50, z-score = -2.22, R) >droSim1.chr3R 10759118 105 - 27517382 ----ACACACACACACCGC-GUGCAUCCUGCGAACCGCAGGAC-UUAUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGUGCCCGUGUAU ----((((.((((.((...-(((..(((((((...))))))).-..(((((.(((((((........))))))).....))))).....)))...)).))))...)))).. ( -30.50, z-score = -2.18, R) >droSec1.super_0 3801178 103 - 21120651 ------ACACACACACCGC-GUGCAUCCUGCGAACCGCAGGAC-UUAUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGUGCCCGUGUAU ------......((((.(.-(..(((((((((...))))))).-..(((((.(((((((........))))))).....)))))..............))..)).)))).. ( -29.80, z-score = -2.12, R) >droYak2.chr3R 8597818 103 + 28832112 ----ACACACACACACCGC-GUGCAUCCUGCGAACCGCAGGAC-UUAUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGCGCUCGUAU-- ----............((.-((((((((((((...))))))).-..(((((.(((((((........))))))).....)))))..............))))).))...-- ( -29.40, z-score = -2.50, R) >droEre2.scaffold_4770 696630 105 - 17746568 ----ACACACACACACUGC-GUGCAUCCUGCGAACCGCAGGAC-UUAUGGCCAUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGCGCUCGUGUAU ----........((((.(.-((((((((((((...))))))).-..(((((.(((((((........))))))).....)))))..............))))).))))).. ( -31.40, z-score = -3.09, R) >droAna3.scaffold_13340 10950570 96 + 23697760 ------------GCA-UGC-GUGCAUCCUGCGAACCGCAGGAC-UCAUGGCCGUGAGCUUUAAUUUCGGUUCAUUUCUCGUUAUCAAAACACUGUGUUUAUGCGCCUGUAU ------------(((-.(.-((((((((((((...))))))).-.(((((..(((((((........))))))).....(((.....))).)))))....))))))))).. ( -25.90, z-score = -0.78, R) >dp4.chr2 27187247 111 + 30794189 AUAUUUAGGUAUAGAUGGUUGUGCAUCCUGCUAACCGCAGGACCUCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGCUAUCAAAACAUUAUGUUUAUGCGCCUGUAU .....(((((...((.(((..((..((((((.....))))))...))..)))(((((((........)))))))...))((.....((((.....))))..)))))))... ( -33.80, z-score = -2.73, R) >droPer1.super_6 2478248 111 + 6141320 AUAUUUAGGUAUAGAUGGUUGUGCAUCCUGCUAACCGCAGGACCUCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGCUAUCAAAACAUUAUGUUUAUGCGCCUGUAU .....(((((...((.(((..((..((((((.....))))))...))..)))(((((((........)))))))...))((.....((((.....))))..)))))))... ( -33.80, z-score = -2.73, R) >droWil1.scaffold_181089 4537936 107 + 12369635 ----GUAUGUAUGUAUAGUCCUGCAUCCUGUUCAACGCAGGAACUCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGUUAUCAAAACAUUAUGUUUAUGUACGUGUAU ----(((((((..(((((((.((..((((((.....))))))...)).))).(((((((........)))))))..................))))..)))))))...... ( -25.70, z-score = -1.78, R) >droVir3.scaffold_13047 2191730 86 - 19223366 ------------------------GCACAUUAACAUGUGUCCU-GCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGUUAUCAAAACAUGGGGCAUAUGCGCGUGUAU ------------------------((((.....((((((((((-........(((((((........))))))).....(((.....)))..))))))))))...)))).. ( -25.20, z-score = -1.42, R) >droMoj3.scaffold_6540 17481041 108 + 34148556 --ACACGUUAGCAUCCUGCAUCGUGUGCGGCAGUCGGGACAAC-GCAUUGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGUUAUCAAAACAUUAGGCCUAUGCGCAUGUAU --..((((..((((...((...((((((((((((((......)-).))))))))(((((........)))))................))))...))..))))..)))).. ( -30.80, z-score = -0.89, R) >consensus ____ACACACACACACCGC_GUGCAUCCUGCGAACCGCAGGAC_UCAUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUAUGCGCGUGUAU ......................(((((((((.....))))))....(((((.(((((((........))))))).....)))))................)))........ (-17.17 = -17.64 + 0.47)
| Location | 4,536,831 – 4,536,942 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.03 |
| Shannon entropy | 0.59572 |
| G+C content | 0.39854 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.36 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4536831 111 + 27905053 --ACCGCAGGACUU-AUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGUGCUCGUGUAUUUGUUUUUAUUCCACUCAUCAAGAUACGGACAUG---- --.(((..(((...-(((((.(((((((........))))))).....)))))((((((((...((....))..)))).)))).......)))..((.....))..))).....---- ( -20.30, z-score = 0.08, R) >droSim1.chr3R 10759146 115 - 27517382 --ACCGCAGGACUU-AUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGUGCCCGUGUAUUUGUUUUUAUUCCACUCAUCGAGAUACGCACAUGCUUA --...(((((....-(((((.(((((((........))))))).....))))).....(((.......))).))((((((((...................))))))))...)))... ( -22.91, z-score = -0.33, R) >droYak2.chr3R 8597846 113 + 28832112 --ACCGCAGGACUU-AUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGCGCUCGU--AUUUGUUUUUAUUCCACUCAUCGGGACGCGCACAUACAAA --...((.((.(..-..).))(((((((........))))))).....))...........(((((.((((((((.--((..((.........))..)))))).)))).))))).... ( -23.70, z-score = -0.53, R) >droEre2.scaffold_4770 696658 115 - 17746568 --ACCGCAGGACUU-AUGGCCAUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUGCGCUCGUGUAUUUGUUUUUAUUCCACUCAUCGGGAUACUCACAUACAUA --..(((.......-(((((.(((((((........))))))).....)))))..((((.....)))))))...((((((.((....((((((.......))))))..)).)))))). ( -24.80, z-score = -2.02, R) >droAna3.scaffold_13340 10950589 108 + 23697760 --ACCGCAGGACUC-AUGGCCGUGAGCUUUAAUUUCGGUUCAUUUCUCGUUAUCAAAACACUGUGUUUAUGCGCCUGUAUUUGUUUUUAUUCCAUUC-UGUGUGUGUGUACG------ --(((((((((...-.(((..(((((((........)))))))..........(((((((..((((....)))).))).))))........))))))-)))).)).......------ ( -19.60, z-score = 0.39, R) >droPer1.super_6 2478281 116 + 6141320 --ACCGCAGGACCUCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGCUAUCAAAACAUUAUGUUUAUGCGCCUGUAUUUGUUUUUGUUCCACUCAUAUAUACAUACAUAUGGAUA --.(((((((.((....))))(((((((........)))))))............((((.....)))).)))...(((((.(((...((.......))...))).)))))...))... ( -22.20, z-score = -0.47, R) >droVir3.scaffold_13047 2191748 88 - 19223366 ------------UGCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGUUAUCAAAACAUGGGGCAUAUGCGCGUGUAUUUGUUGUUUUAGUUUUUAUU------------------ ------------.(((((((((((((((........))))))).....(((.....)))....))).)))))............................------------------ ( -20.00, z-score = -0.88, R) >droMoj3.scaffold_6540 17481069 95 + 34148556 CAGUCGGGACAACGCAUUGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGUUAUCAAAACAUUAGGCCUAUGCGCAUGUAUUUGUUGUUUUUUAUU----------------------- (((.(((((((.(((((.((((((((((........))))))).....(((.....)))....)))..)))))..))).))))))).........----------------------- ( -22.10, z-score = -1.36, R) >droGri2.scaffold_14906 10677335 100 - 14172833 ACGGCAGGACAACGCAUGGCCGUGAGCUUUAAUUUUGGCUCAUUUUUCGUUAUCAAAACAUUUGGCUUAUGUGCGUGUACUUGUUGUUAUUGUUUUUUAC------------------ (((((((((((.((((((((((((((((........))))))).....(((.....)))....))))...)))))))).)))))))).............------------------ ( -31.90, z-score = -3.83, R) >consensus __ACCGCAGGACUU_AUGGCCGUGAGCUUUAAUUUCGGCUCAUUUUUCGCUAUCAAAACACUAUGUUUAUGCGCGUGUAUUUGUUUUUAUUCCACUCAUC__GAUACGCACAU_____ ........(((....(((((.(((((((........))))))).....)))))((((((((...((....))..)))).)))).......)))......................... (-12.41 = -12.36 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:05 2011