| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,525,078 – 4,525,172 |
| Length | 94 |
| Max. P | 0.947782 |

| Location | 4,525,078 – 4,525,172 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.83 |
| Shannon entropy | 0.60416 |
| G+C content | 0.53235 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.78 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
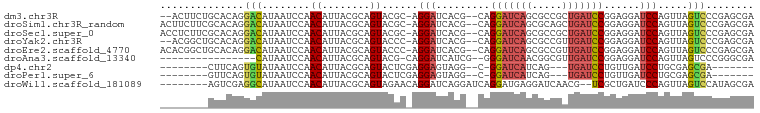
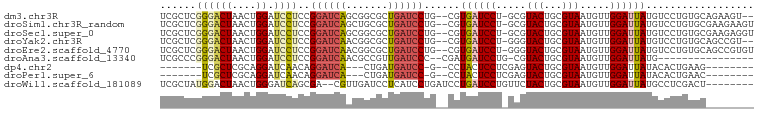
>dm3.chr3R 4525078 94 + 27905053 --ACUUCUGCACAGGACAUAAUCCAACAUUACGCAGUACGC-AGGAUCACG--CAGGAUCAGCGCCGCUGAUCCGGAGGAUCCAGUUAGUCCCGAGCGA --.....(((.(.((((...............((.....))-.(((((.(.--..((((((((...))))))))...)))))).....)))).).))). ( -32.80, z-score = -2.97, R) >droSim1.chr3R_random 295384 96 + 1307089 ACUUCUUCGCACAGGACAUAAUCCAACAUUACGCAGUACGC-AGGAUCACG--CAGGAUCAGCGCAGCUGAUCCGGAGGAUCCAGUUAGUCCCGAGCGA ......((((.(.((((...............((.....))-.(((((.(.--..((((((((...))))))))...)))))).....)))).).)))) ( -35.50, z-score = -4.24, R) >droSec1.super_0 3791929 96 - 21120651 ACCUCUUCGCACAGGACAUAAUCCAACAUUACGCAGUACGC-AGGAUCACG--CAGGAUCAGCGCCGCUGAUCCGGAGGAUCCAGUUAGUCCCGAGCGA ......((((.(.((((...............((.....))-.(((((.(.--..((((((((...))))))))...)))))).....)))).).)))) ( -35.50, z-score = -3.73, R) >droYak2.chr3R 8588114 94 + 28832112 --ACGGCUGCACAGGACAUAAUCCAACAUUACGCAGUACCC-AGGAUCACG--CAGGAUCAGCGCCGUUGAUCCGGAGGAUCCAGUUAGUCCCGAGCGA --.....(((.(.((((........((........))....-.(((((.(.--..((((((((...))))))))...)))))).....)))).).))). ( -27.70, z-score = -1.24, R) >droEre2.scaffold_4770 685795 96 - 17746568 ACACGGCUGCACAGGACAUAAUCCAACAUUACGCAGUACCC-AGGAUCACG--CAGGAUCAGCGCCGUUGAUCCGGAGGAUCCAGUUAGUCCCGAGCGA .......(((.(.((((........((........))....-.(((((.(.--..((((((((...))))))))...)))))).....)))).).))). ( -27.70, z-score = -1.28, R) >droAna3.scaffold_13340 10942677 80 + 23697760 ----------------CAUAAUCCAACAUUACGCAGUACG-CAGGAUCAUCG--GGGAUCAACGGCGUUGAUCCGGAGGAUCCAGUUAGUCCCGGGCGA ----------------.....(((........((.....)-).(((((.((.--.((((((((...)))))))).)).)))))..........)))... ( -22.70, z-score = -0.47, R) >dp4.chr2 27178112 78 + 30794189 --------CUUCAGUGUAUAAUCCAACAUUACGCAGUACUCGAGGAGUAGG--C-GGAUCAUCAG---UGAUCCUGUUGAUCCUGCGAGCGA------- --------.....(((((...........)))))....((((((((...((--(-((((((....---)))))).)))..)))).))))...------- ( -24.40, z-score = -1.75, R) >droPer1.super_6 2469067 78 + 6141320 --------GUUCAGUGUAUAAUCCAACAUUACGCAGUACUCGAGGAGUAGG--C-GGAUCAUCAG---UGAUCCUGUUGAUCCUGCGAGCGA------- --------.....(((((...........)))))....((((((((...((--(-((((((....---)))))).)))..)))).))))...------- ( -24.40, z-score = -1.75, R) >droWil1.scaffold_181089 4529973 89 + 12369635 --------AGUCGAGGCAUAAUCCAACAUUACGCAGUAGAACAGGAUCAGGAUCAGGAUGAGGAUCAACG--UCGCUGAUCCCAGUUAGUCCAUAGCGA --------.((....))..............(((.((.((....(((..(((((((((((........))--)).)))))))..)))..)).)).))). ( -22.10, z-score = -1.21, R) >consensus ________GCACAGGACAUAAUCCAACAUUACGCAGUACGC_AGGAUCACG__CAGGAUCAGCGCCGCUGAUCCGGAGGAUCCAGUUAGUCCCGAGCGA ..............(((....(((.....(((...))).....))).........(((((((.....)))))))..............)))........ (-10.85 = -10.78 + -0.07)
| Location | 4,525,078 – 4,525,172 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.83 |
| Shannon entropy | 0.60416 |
| G+C content | 0.53235 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
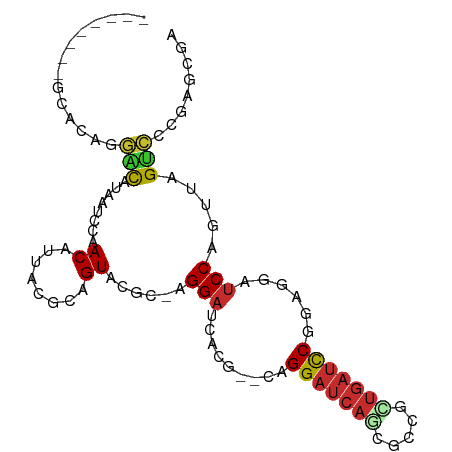
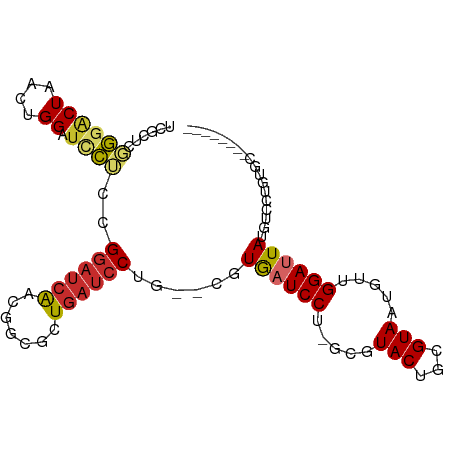
>dm3.chr3R 4525078 94 - 27905053 UCGCUCGGGACUAACUGGAUCCUCCGGAUCAGCGGCGCUGAUCCUG--CGUGAUCCU-GCGUACUGCGUAAUGUUGGAUUAUGUCCUGUGCAGAAGU-- ..((.((((((.....((.....))((((((((...))))))))..--.(((((((.-((((........)))).))))))))))))).))......-- ( -37.50, z-score = -2.74, R) >droSim1.chr3R_random 295384 96 - 1307089 UCGCUCGGGACUAACUGGAUCCUCCGGAUCAGCUGCGCUGAUCCUG--CGUGAUCCU-GCGUACUGCGUAAUGUUGGAUUAUGUCCUGUGCGAAGAAGU ((((.((((((.....((.....))((((((((...))))))))..--.(((((((.-((((........)))).))))))))))))).))))...... ( -40.60, z-score = -4.29, R) >droSec1.super_0 3791929 96 + 21120651 UCGCUCGGGACUAACUGGAUCCUCCGGAUCAGCGGCGCUGAUCCUG--CGUGAUCCU-GCGUACUGCGUAAUGUUGGAUUAUGUCCUGUGCGAAGAGGU ((((.((((((.....((.....))((((((((...))))))))..--.(((((((.-((((........)))).))))))))))))).))))...... ( -40.60, z-score = -3.18, R) >droYak2.chr3R 8588114 94 - 28832112 UCGCUCGGGACUAACUGGAUCCUCCGGAUCAACGGCGCUGAUCCUG--CGUGAUCCU-GGGUACUGCGUAAUGUUGGAUUAUGUCCUGUGCAGCCGU-- ..((.((((((...((((.....)))).(((((.((((..((((..--.........-))))...))))...))))).....)))))).))......-- ( -32.30, z-score = -1.02, R) >droEre2.scaffold_4770 685795 96 + 17746568 UCGCUCGGGACUAACUGGAUCCUCCGGAUCAACGGCGCUGAUCCUG--CGUGAUCCU-GGGUACUGCGUAAUGUUGGAUUAUGUCCUGUGCAGCCGUGU ..((.((((((...((((.....)))).(((((.((((..((((..--.........-))))...))))...))))).....)))))).))........ ( -32.30, z-score = -0.81, R) >droAna3.scaffold_13340 10942677 80 - 23697760 UCGCCCGGGACUAACUGGAUCCUCCGGAUCAACGCCGUUGAUCCC--CGAUGAUCCUG-CGUACUGCGUAAUGUUGGAUUAUG---------------- .....(((........((.....))((((((((...)))))))))--))(((((((.(-(((........)))).))))))).---------------- ( -25.30, z-score = -1.81, R) >dp4.chr2 27178112 78 - 30794189 -------UCGCUCGCAGGAUCAACAGGAUCA---CUGAUGAUCC-G--CCUACUCCUCGAGUACUGCGUAAUGUUGGAUUAUACACUGAAG-------- -------..(((((.((((......((((((---....))))))-.--.....)))))))))..((..((((.....))))..))......-------- ( -20.90, z-score = -1.38, R) >droPer1.super_6 2469067 78 - 6141320 -------UCGCUCGCAGGAUCAACAGGAUCA---CUGAUGAUCC-G--CCUACUCCUCGAGUACUGCGUAAUGUUGGAUUAUACACUGAAC-------- -------..(((((.((((......((((((---....))))))-.--.....)))))))))..((..((((.....))))..))......-------- ( -20.90, z-score = -1.56, R) >droWil1.scaffold_181089 4529973 89 - 12369635 UCGCUAUGGACUAACUGGGAUCAGCGA--CGUUGAUCCUCAUCCUGAUCCUGAUCCUGUUCUACUGCGUAAUGUUGGAUUAUGCCUCGACU-------- .(((...((((.....((((((((.((--.(........).))))))))))......))))....)))....(((((........))))).-------- ( -21.80, z-score = -1.03, R) >consensus UCGCUCGGGACUAACUGGAUCCUCCGGAUCAACGGCGCUGAUCCUG__CGUGAUCCU_GCGUACUGCGUAAUGUUGGAUUAUGUCCUGUGC________ ......((((((....)).))))..((((((.......))))))......((((((.....(((...))).....)))))).................. (-11.48 = -11.72 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:59:00 2011