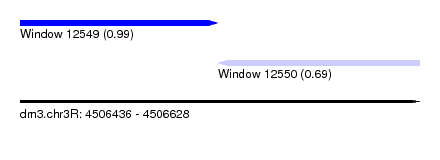
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,506,436 – 4,506,628 |
| Length | 192 |
| Max. P | 0.994541 |

| Location | 4,506,436 – 4,506,531 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 63.01 |
| Shannon entropy | 0.68514 |
| G+C content | 0.36529 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -8.80 |
| Energy contribution | -10.08 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
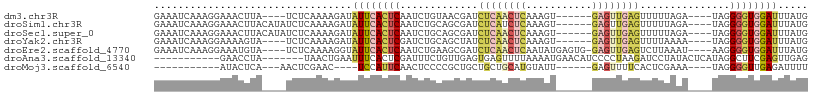
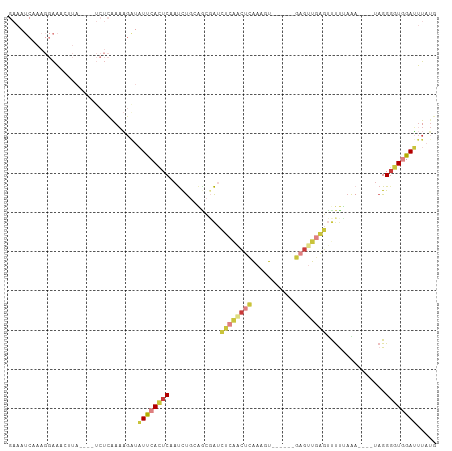
>dm3.chr3R 4506436 95 + 27905053 GAAAUCAAAGGAAACUUA----UCUCAAAAGAUAUUCACUCAAUCUGUAACGAUCUCAACUCAAAGU------GAGUUGAGUUUUUAGA----UAGGGGUGGAUUUAUG .(((((.(((....)))(----(((....))))...(((((.(((((.......((((((((.....------))))))))....))))----)..))))))))))... ( -27.00, z-score = -3.47, R) >droSim1.chr3R 10738819 99 - 27517382 GAAAUCAAAGGAAACUUACAUAUCUCAAAAGAUAUUCACUCAAUCUGCAGCGAUCUCAUCUCAAAGU------GAGUUGAGUUUUUAGA----UAGGGGUGGAUUUAUG .(((((.(((....)))..((((((....)))))).(((((.(((((.......((((.(((.....------))).))))....))))----)..))))))))))... ( -24.10, z-score = -2.03, R) >droSec1.super_0 3773430 99 - 21120651 GAAAUCAAAGGAAACUUACAUAUCUCAAAAGAUAUUCACUCAAUCUGCAGCGAUCUCAACUCAAAGU------GAGUUGAGUUUUUAGA----UAGGGGUGGAUUUAUG .(((((.(((....)))..((((((....)))))).(((((.(((((.......((((((((.....------))))))))....))))----)..))))))))))... ( -28.20, z-score = -3.64, R) >droYak2.chr3R 8569302 95 + 28832112 GAAAUCAAAGGAAAAGUA----UCUCAAAAGAUAUUCACUCGAUCUGCAGCUAUCUCAACUCAAAGU------GAGUUGAGUUUUAAAA----UAGGGGUGGAUUUAUG ..........((..((((----(((....)))))))...))((((..(..((((((((((((.....------)))))))).......)----)))..)..)))).... ( -22.81, z-score = -2.39, R) >droEre2.scaffold_4770 666765 100 - 17746568 GAAAUCAAAGGAAAUGUA----UCUCAAAAGGUAUUCACUCAAUCUGAAGCGAUCUCAACUCAAUAUGAGUG-GAGUUGAGUCUUAAAU----AAGGGGUGGAUUUAUG .(((((.........(((----(((....)))))).(((((......(((....((((((((.((....)).-)))))))).)))....----...))))))))))... ( -22.42, z-score = -1.49, R) >droAna3.scaffold_13340 10923275 91 + 23697760 -----------GAACCUA-------UAACUGAAUUUCACUCGAUUUCUGUUGAGUGAGUUUUAAAAUGAACAUCCCCUAAGAUCCUAUACUCAUAGGCUUCGAGUUGAG -----------.......-------..........((((((((..(((((.(((((.((((......))))(((......)))....))))))))))..))))).))). ( -20.00, z-score = -2.16, R) >droMoj3.scaffold_6540 17444908 81 + 34148556 -----------AUACUCA---AACUCGAAC----UCCAUUCAACUCCCCGCUGCUGCUGCAUGUAUU------GAGUUUUCACUCGAAA----UAGGGGUUGAGAUUUU -----------...((((---(....(((.----....)))....(((((((((....))).)).((------((((....))))))..----..)))))))))..... ( -20.40, z-score = -2.16, R) >consensus GAAAUCAAAGGAAACUUA____UCUCAAAAGAUAUUCACUCAAUCUGCAGCGAUCUCAACUCAAAGU______GAGUUGAGUUUUUAAA____UAGGGGUGGAUUUAUG .................................((((((((.............((((((((...........))))))))...............))))))))..... ( -8.80 = -10.08 + 1.27)
| Location | 4,506,531 – 4,506,628 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Shannon entropy | 0.28709 |
| G+C content | 0.34310 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.28 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
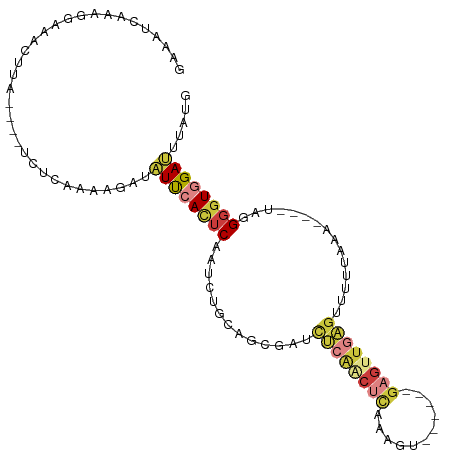
>dm3.chr3R 4506531 97 - 27905053 AGUGGAUGUUGUUCUUUCUUUGGCCACUCAAAAAGUGAGUAAAAGAGAGUGCUACUCAAAACUAAGAAAAUCUAUAUCUCGAUGGAUCCUCAUACAA (((.((.((.((.((((((((.((((((.....)))).)).)))))))).)).))))...)))..((..((((((......))))))..))...... ( -23.10, z-score = -1.84, R) >droSim1.chr3R 10738918 90 + 27517382 -------AGUGGAUUUUUUUUUGCCACUCAAAAAGUGAGUAAAAGAGAGUGCAACUCAAAAUUGAGAAAAUCUAUAGUUCACUGGAUCCUCAUAAAA -------..((.((((((((((((((((.....)))).)))))))))))).)).........((((...(((((........))))).))))..... ( -20.90, z-score = -2.01, R) >droSec1.super_0 3773529 89 + 21120651 -------AGUGGA-UUUUUUUUACCACUCAAAAAGUGAGUAAAAGAGAGUGCUACUCAAAAUUGAGAAAAUCUUUAGCUCGCUGGAUCCUCAUAAAA -------(((((.-((((((((((((((.....)))).))))))))))...)))))......((((...((((..((....)))))).))))..... ( -20.50, z-score = -1.70, R) >droYak2.chr3R 8569397 88 - 28832112 -------AGUGGAUUUCCUUUGCCCACUCAAAAAGUGAGUAAAAGAGAGUUAUACUCAAAAC--AAAAAAUCUAUAGCUCACUGGAUCCUCAAAGAA -------.(.(((((((.(((((.((((.....)))).))))).))((((((((........--........))))))))....))))).)...... ( -19.89, z-score = -2.41, R) >consensus _______AGUGGAUUUUCUUUGGCCACUCAAAAAGUGAGUAAAAGAGAGUGCUACUCAAAACUGAGAAAAUCUAUAGCUCACUGGAUCCUCAUAAAA .......((((.....((((((((((((.....)))).))))))))(((.....)))......................)))).............. (-13.46 = -13.28 + -0.19)

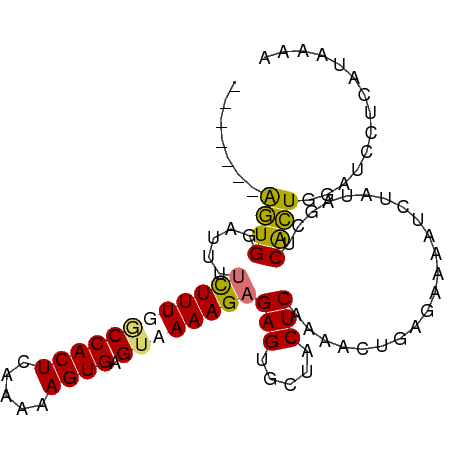
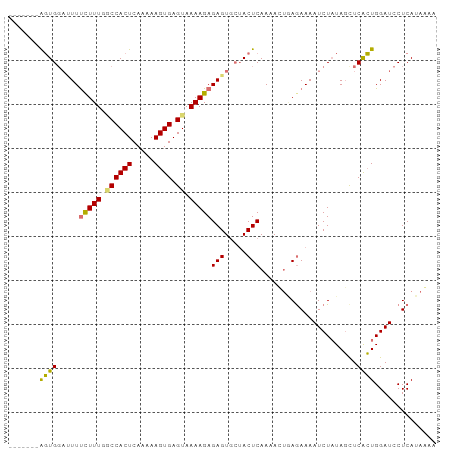
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:55 2011