| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,769,229 – 6,769,327 |
| Length | 98 |
| Max. P | 0.999866 |

| Location | 6,769,229 – 6,769,327 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 67.27 |
| Shannon entropy | 0.43694 |
| G+C content | 0.36904 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
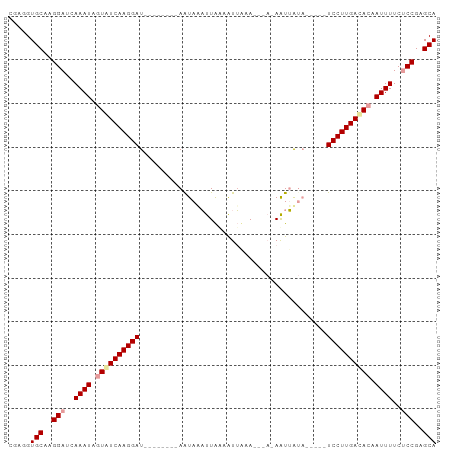
>dm3.chr2L 6769229 98 + 23011544 CGAAGUGCAAGGAUCAAAUACUAUCAAGGAUCAAUGAUUAAUAAAUUAACAUUUCAUUUACAAUUAUAACUGAUCCUUGACACAAUUUUCGCCGAGCA .....(((..((...((((....((((((((((((((((..(((((.........))))).))))))...))))))))))....))))...))..))) ( -19.90, z-score = -3.05, R) >droEre2.scaffold_4929 15685410 78 + 26641161 CGAGGUGCUCGGAUCAAAUGGUGUCAAGGAU--------AAUAUCCUGCAGUGCUA-----AGUGA-------UCCUUGACACAAUUUUCUCCGAGCA .....((((((((..((((.(((((((((((--------........((...))..-----....)-------)))))))))).))))..)))))))) ( -32.84, z-score = -5.00, R) >droYak2.chr2L 16184113 82 - 22324452 CGUGGUGCAAGGAUCAAAUAGUAUCAAGGAU--------AGUACAUUAUAAUUAAA---AGAAUUGUA-----UCCUUGAUACAAUUUUCUCCGAGCA .....(((..(((..((((.((((((((((.--------.......(((((((...---..)))))))-----)))))))))).))))..)))..))) ( -23.60, z-score = -4.42, R) >consensus CGAGGUGCAAGGAUCAAAUAGUAUCAAGGAU________AAUAAAUUAAAAUUAAA___A_AAUUAUA_____UCCUUGACACAAUUUUCUCCGAGCA .....(((..(((..((((.((((((((((...........................................)))))))))).))))..)))..))) (-16.18 = -16.96 + 0.78)


| Location | 6,769,229 – 6,769,327 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 67.27 |
| Shannon entropy | 0.43694 |
| G+C content | 0.36904 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -5.57 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.63 |
| SVM RNA-class probability | 0.999866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
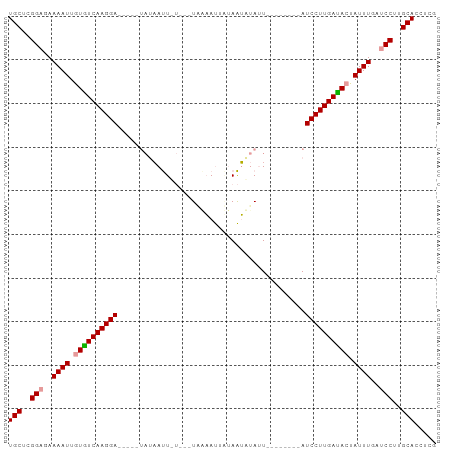
>dm3.chr2L 6769229 98 - 23011544 UGCUCGGCGAAAAUUGUGUCAAGGAUCAGUUAUAAUUGUAAAUGAAAUGUUAAUUUAUUAAUCAUUGAUCCUUGAUAGUAUUUGAUCCUUGCACUUCG (((..((...((((..((((((((((((((.((....((((((.........))))))..)).))))))))))))))..))))...))..)))..... ( -26.90, z-score = -3.91, R) >droEre2.scaffold_4929 15685410 78 - 26641161 UGCUCGGAGAAAAUUGUGUCAAGGA-------UCACU-----UAGCACUGCAGGAUAUU--------AUCCUUGACACCAUUUGAUCCGAGCACCUCG ((((((((..((((.((((((((((-------(..((-----(.((...))))).....--------))))))))))).))))..))))))))..... ( -34.50, z-score = -6.70, R) >droYak2.chr2L 16184113 82 + 22324452 UGCUCGGAGAAAAUUGUAUCAAGGA-----UACAAUUCU---UUUAAUUAUAAUGUACU--------AUCCUUGAUACUAUUUGAUCCUUGCACCACG (((..(((..((((.((((((((((-----((.......---................)--------))))))))))).))))..)))..)))..... ( -25.00, z-score = -6.09, R) >consensus UGCUCGGAGAAAAUUGUGUCAAGGA_____UAUAAUU_U___UAAAAUUAUAAUAUAUU________AUCCUUGAUACUAUUUGAUCCUUGCACCUCG (((..(((..((((.((((((((((...........................................)))))))))).))))..)))..)))..... (-19.04 = -19.26 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:56 2011