
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,480,206 – 4,480,350 |
| Length | 144 |
| Max. P | 0.694124 |

| Location | 4,480,206 – 4,480,350 |
|---|---|
| Length | 144 |
| Sequences | 6 |
| Columns | 161 |
| Reading direction | forward |
| Mean pairwise identity | 77.15 |
| Shannon entropy | 0.40547 |
| G+C content | 0.37736 |
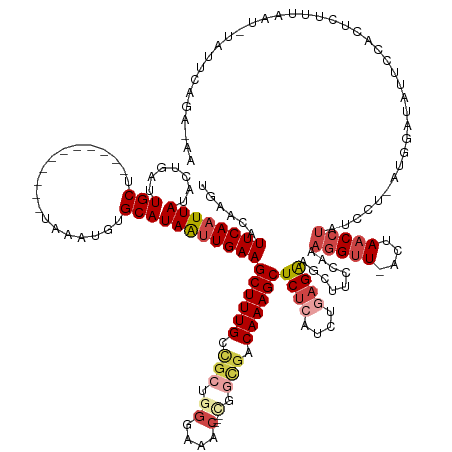
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -14.90 |
| Energy contribution | -16.95 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.694124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
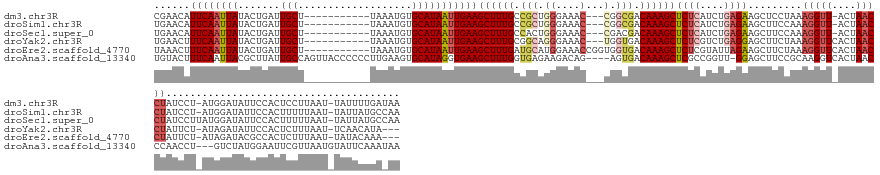
>dm3.chr3R 4480206 144 + 27905053 CGAACAUUCAAUUAUACUGAUUGCU-----------UAAAUGUGCAUAAUUGAAGCUUUGCCGCUGGGAAAC---CGGCGACAAAGCUCUCAUCUGAGAAGCUCCUAAAGGUU-ACUAACCUAUCCU-AUGGAUAUUCCACUCCUUAAU-UAUUUUGAUAA ......((((((((.......(((.-----------.......)))))))))))((((((.((((((....)---))))).))))))((((....))))........((((..-.......(((((.-..))))).......))))...-........... ( -38.00, z-score = -3.75, R) >droSim1.chr3R 10712547 144 - 27517382 UGAACAUUCAAUUAUACUGAUUGCU-----------UAAAUGUGCAUAAUUGAAGCUUUGCCGCUGGGAAAC---CGGCGACAAAGCUCUCAUCUGAGAAGCUUCCAAAGGUU-ACUAACCUAUCCU-AUGGAUAUUCCACUUUUUAAU-UAUUAUGCCAA ...((((((((((.....)))))..-----------..)))))(((((((((((((((((.((((((....)---))))).))))))).)))..((.(((...((((.(((.(-(......)).)))-.))))..))))).........-.)))))))... ( -44.00, z-score = -5.27, R) >droSec1.super_0 3747257 145 - 21120651 UGAACAUUCAAUUAUACUGAUUGCU-----------UAAAUGUGCAUAAUUGAAGCUUUGCCACUGGGAAAC---CGACGACAAAGCUCUCAUCUGAGAAGCUUCCAAAGGUU-ACUAACCUAUCCUUAUGGAUAUUCCACUUUUUAAU-UAUUAUGCCAA ...((((((((((.....)))))..-----------..)))))(((((((((((((((((.(..(((....)---))..).))))))).)))..(((((((.......((((.-....))))((((....))))......)))))))..-.)))))))... ( -34.50, z-score = -3.30, R) >droYak2.chr3R 8543081 142 + 28832112 UGAACUUUCAAUUAUACUGAUUGCU-----------UAAAUGUGCAUAAUUGAAGCUUUGCGGCAGGGAAAC---UGGUGACAAAGCUCUCGUCUGAGGAGCUUCUAAAGGUUCACUAACCUAUUCU-AUAGAUAUUCCACUCUUUAAU-UCAACAUA--- ((((..((((((((.......(((.-----------.......)))))))))))((((((((.(((.....)---)).)).))))))..........((((..((((.(((((....))))).....-.))))..)))).........)-))).....--- ( -30.61, z-score = -0.85, R) >droEre2.scaffold_4770 640224 145 - 17746568 UAAACUUUCAAUUAUACUGAUUGCU-----------UAAAUGUGCAUAAUUGAAGCUUUGAUGCAUGGAAACCGGUGGUGACAAAGCUCUCGUAUUAGAAGCUUCUAAAGGUUCACUAACCUAUUCU-AUAGAUACGCCACUCUUUAAU-UAUACAAA--- ........(((((.....)))))..-----------......((.((((((((((...........(....).(((((((...(((((.((......)))))))....(((((....))))).....-.......))))))))))))))-))).))..--- ( -28.00, z-score = -0.87, R) >droAna3.scaffold_13340 18717038 153 + 23697760 UGUACUUUCAAUUACGCUUAUUGCCAGUUACCCCCCUUGAAGUGCAUAGGUGAAGCUUUGGUGAGAAGACAG----AGUGACAAAGCUCGCCGGUU-GGAGCUUCCGCAAGGUCACUAACCCAACCU---GUCUAUGGAAUUCGUUAAUGUAUUCAAAUAA .(((((..((((.......))))..)).))).....(((((.(((((.(((((.((((((............----.....)))))))))))((((-((((...((....))...))...)))))).---.................)))))))))).... ( -36.33, z-score = 0.19, R) >consensus UGAACAUUCAAUUAUACUGAUUGCU___________UAAAUGUGCAUAAUUGAAGCUUUGCCGCUGGGAAAC___CGGCGACAAAGCUCUCAUCUGAGAAGCUUCCAAAGGUU_ACUAACCUAUCCU_AUGGAUAUUCCACUCUUUAAU_UAUUCAGA_AA ......((((((((.......(((...................)))))))))))((((((.(((..(....).....))).)))))).......(((((((.......(((((....)))))((((....))))......))))))).............. (-14.90 = -16.95 + 2.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:51 2011