| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,416,376 – 4,416,453 |
| Length | 77 |
| Max. P | 0.937690 |

| Location | 4,416,376 – 4,416,453 |
|---|---|
| Length | 77 |
| Sequences | 8 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.42401 |
| G+C content | 0.41146 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -13.27 |
| Energy contribution | -13.65 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
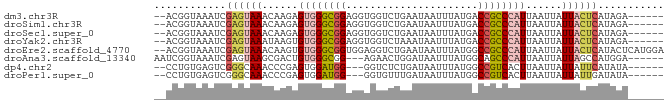
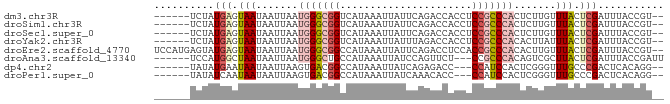
>dm3.chr3R 4416376 77 + 27905053 --ACGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAUAGA------ --..........((((((.....(((((((((..((((..........))))..))))))))).....)))))).....------ ( -23.90, z-score = -3.28, R) >droSim1.chr3R 10656471 77 - 27517382 --ACGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAUAGA------ --..........((((((.....(((((((((..((((..........))))..))))))))).....)))))).....------ ( -23.90, z-score = -3.28, R) >droSec1.super_0 3692115 77 - 21120651 --ACGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAUAGA------ --..........((((((.....(((((((((..((((..........))))..))))))))).....)))))).....------ ( -23.90, z-score = -3.28, R) >droYak2.chr3R 8485548 77 + 28832112 --ACGGUAAAUCGAGUAAAUAAGUGUGGGCGGAGGUGGUCUAAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAUAGA------ --..........((((((.(((..((((((((..((((..........))))..))))))))...))))))))).....------ ( -23.70, z-score = -3.31, R) >droEre2.scaffold_4770 582727 83 - 17746568 --ACGGUAAAUCGAGUAAACAAGUGUGGGCGGUGGAGGUCUGAAUAAUUUAUGGCCGCCCAUUAAUUAUUACUCAUACUCAUGGA --..........((((((......(((((((((.(((((.......)))).).)))))))))......))))))........... ( -23.90, z-score = -1.86, R) >droAna3.scaffold_13340 18658890 76 + 23697760 AAUCGGUAAAUCGAGUAAGCGACUGUGGGCGG---AGAACUGGAUAAUUUAUGGCAGCCCAUUAAUUAUUAGCCAUGGA------ ..((((....))))........((((((.(((---....)))(((((((.((((....)))).)))))))..)))))).------ ( -16.20, z-score = 0.06, R) >dp4.chr2 13526931 74 - 30794189 --CCUGUGAGUCGGGCAAACCCGAGUGGAUGG---GGUCUCUGAUAAUUUAUGGCCGUCACUUAAUUAUUAUUCAUAUA------ --..(((((((.(((....)))((((((....---((((..(((....))).)))).)))))).......)))))))..------ ( -17.80, z-score = -0.22, R) >droPer1.super_0 711954 74 - 11822988 --CCUGUGAGUCGGGCAAACCCGAGUGGAUGG---GGUGUUUGAUAAUUUAUGGCCGUCACUUAAUUAUUAUUGAUAUA------ --(((((.(.(((((....))))).)..))))---)(((((((((((((...((......)).))))))))..))))).------ ( -16.00, z-score = 0.19, R) >consensus __ACGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAUAGA______ ............((((((......((((((((......................))))))))......))))))........... (-13.27 = -13.65 + 0.38)
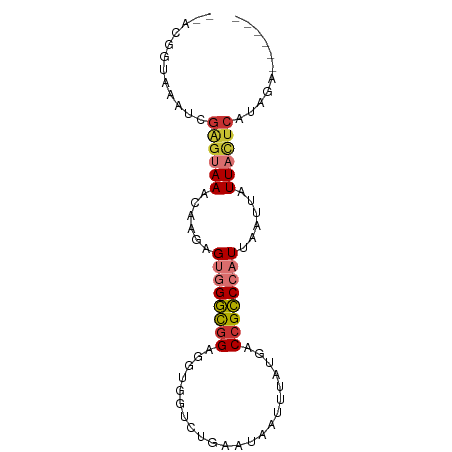
| Location | 4,416,376 – 4,416,453 |
|---|---|
| Length | 77 |
| Sequences | 8 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.42401 |
| G+C content | 0.41146 |
| Mean single sequence MFE | -16.22 |
| Consensus MFE | -7.49 |
| Energy contribution | -8.10 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
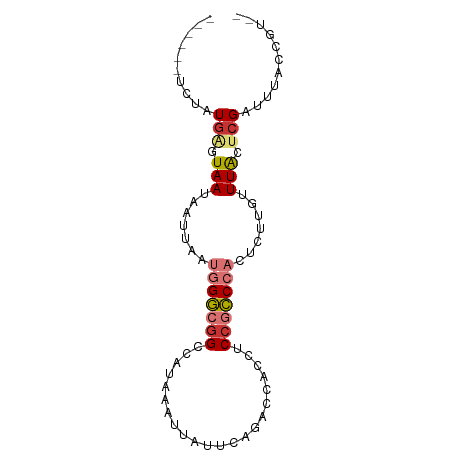
>dm3.chr3R 4416376 77 - 27905053 ------UCUAUGAGUAAUAAUUAAUGGGCGGUCAUAAAUUAUUCAGACCACCUCCGCCCACUCUUGUUUACUCGAUUUACCGU-- ------....(((((((.......(((((((......................))))))).......))))))).........-- ( -19.29, z-score = -3.17, R) >droSim1.chr3R 10656471 77 + 27517382 ------UCUAUGAGUAAUAAUUAAUGGGCGGUCAUAAAUUAUUCAGACCACCUCCGCCCACUCUUGUUUACUCGAUUUACCGU-- ------....(((((((.......(((((((......................))))))).......))))))).........-- ( -19.29, z-score = -3.17, R) >droSec1.super_0 3692115 77 + 21120651 ------UCUAUGAGUAAUAAUUAAUGGGCGGUCAUAAAUUAUUCAGACCACCUCCGCCCACUCUUGUUUACUCGAUUUACCGU-- ------....(((((((.......(((((((......................))))))).......))))))).........-- ( -19.29, z-score = -3.17, R) >droYak2.chr3R 8485548 77 - 28832112 ------UCUAUGAGUAAUAAUUAAUGGGCGGUCAUAAAUUAUUUAGACCACCUCCGCCCACACUUAUUUACUCGAUUUACCGU-- ------....(((((((.......(((((((......................))))))).......))))))).........-- ( -19.29, z-score = -3.65, R) >droEre2.scaffold_4770 582727 83 + 17746568 UCCAUGAGUAUGAGUAAUAAUUAAUGGGCGGCCAUAAAUUAUUCAGACCUCCACCGCCCACACUUGUUUACUCGAUUUACCGU-- ....(((((.(((((((.......(((((((......................))))))).......))))))))))))....-- ( -20.39, z-score = -2.60, R) >droAna3.scaffold_13340 18658890 76 - 23697760 ------UCCAUGGCUAAUAAUUAAUGGGCUGCCAUAAAUUAUCCAGUUCU---CCGCCCACAGUCGCUUACUCGAUUUACCGAUU ------....(((...((((((.((((....)))).))))))))).....---........(((((......)))))........ ( -10.00, z-score = 0.70, R) >dp4.chr2 13526931 74 + 30794189 ------UAUAUGAAUAAUAAUUAAGUGACGGCCAUAAAUUAUCAGAGACC---CCAUCCACUCGGGUUUGCCCGACUCACAGG-- ------..................(((((((.((............((((---(.........))))))).)))..))))...-- ( -11.20, z-score = 0.35, R) >droPer1.super_0 711954 74 + 11822988 ------UAUAUCAAUAAUAAUUAAGUGACGGCCAUAAAUUAUCAAACACC---CCAUCCACUCGGGUUUGCCCGACUCACAGG-- ------..................((((.((...................---))......(((((....))))).))))...-- ( -11.01, z-score = -0.52, R) >consensus ______UCUAUGAGUAAUAAUUAAUGGGCGGCCAUAAAUUAUUCAGACCACCUCCGCCCACUCUUGUUUACUCGAUUUACCGU__ ..........(((.(((.......(((((((......................))))))).......))).)))........... ( -7.49 = -8.10 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:47 2011