| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,410,021 – 4,410,131 |
| Length | 110 |
| Max. P | 0.996949 |

| Location | 4,410,021 – 4,410,131 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Shannon entropy | 0.44816 |
| G+C content | 0.48033 |
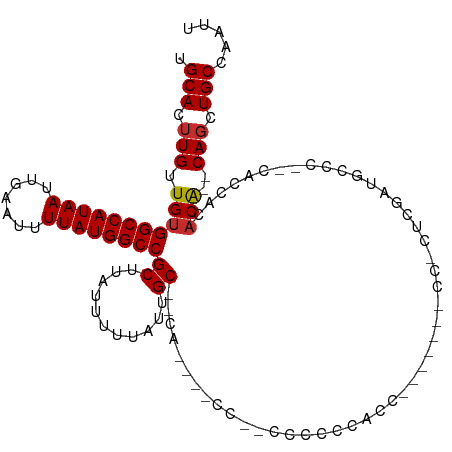
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
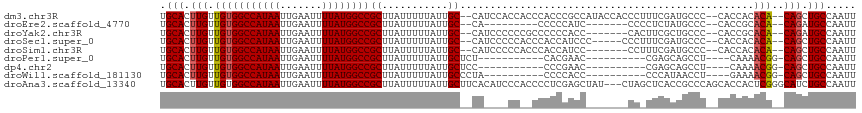
>dm3.chr3R 4410021 110 + 27905053 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGC--CAUCCACCACCCACCCGCCAUACCACCCUUUCGAUGCCC--CACCACACA--CAGCUGCCAAUU .(((.((((.((((((((((.......)))))))))).............--((((............................))))...--........)--))).)))..... ( -20.09, z-score = -1.83, R) >droEre2.scaffold_4770 576553 94 - 17746568 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGC--CA---------CCCCCAUC-------CCCCUCUAUGCCC--CACCGCACA--CAGAUGCCAAUU .(((.((((.((((((((((.......)))))))))).............--..---------........-------........(((..--....))).)--))).)))..... ( -21.80, z-score = -3.00, R) >droYak2.chr3R 8479281 103 + 28832112 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGC--CAUCCCCCCGCCCCCCACC-------CACUUCGCUGCCC--CACCGCACA--CAGAUGCCAAUU .(((.((((.((((((((((.......)))))))))).............--...................-------......(.(((..--....)))))--))).)))..... ( -21.90, z-score = -2.24, R) >droSec1.super_0 3685845 105 - 21120651 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGC--CAUCCCCCACCCACCAUCCC-----CCCUUUCGAUGCCC--CACCACACA--CAGCUGCCAAUU .(((.((((.((((((((((.......)))))))))).............--..............((((..-----.......))))...--........)--))).)))..... ( -21.30, z-score = -2.89, R) >droSim1.chr3R 10650186 103 - 27517382 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGC--CAUCCCCCACCCACCAUCC-------CCUUUCGAUGCCC--CACCACACA--CAGCUGCCAAUU .(((.((((.((((((((((.......)))))))))).............--..............((((.-------......))))...--........)--))).)))..... ( -21.80, z-score = -2.91, R) >droPer1.super_0 705508 90 - 11822988 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGCUCU-----------CACGAAC----------CGAGCAGCCU----CAAAACGG-CAGCUGCCAAUU .(((.(((((((((((((((.......))))))))...........((((((.-----------.......----------.))))))...----....))))-))).)))..... ( -28.00, z-score = -2.72, R) >dp4.chr2 13520495 90 - 30794189 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGCUCC-----------CCCGAAC----------CGAGCAGCCU----CAAAACGG-CAGCUGCCAAUU .(((.(((((((((((((((.......))))))))...........((((((.-----------.......----------.))))))...----....))))-))).)))..... ( -27.50, z-score = -2.70, R) >droWil1.scaffold_181130 7811961 91 - 16660200 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGCCCUA----------CCCCACC----------CCCAUAACCU----GAAAACGG-CAGCUGCCAAUU .(((.(((((((((((((((.......))))))))((...........))....----------.......----------..........----....))))-))).)))..... ( -22.00, z-score = -2.30, R) >droAna3.scaffold_13340 18652630 113 + 23697760 UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGCUUCACAUCCCACCCCUCGAGCUAU---CUAGCUCACCGCCCAGCACCACUCGGGCAUCUGCCAAUU .(((......((((((((((.......))))))))))..............................(((((..---..)))))...((((((.....)).))))...)))..... ( -31.90, z-score = -3.24, R) >consensus UGCACUUGUUGUGGCCAUAAUUGAAUUUUAUGGCCGCUUAUUUUUAUUGC__CA____CC__CCCCCCACC_______CC_CUCGAUGCCC__CACCACACA__CAGCUGCCAAUU .(((.(((..((((((((((.......)))))))))).((....))..........................................................))).)))..... (-15.94 = -16.06 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:46 2011