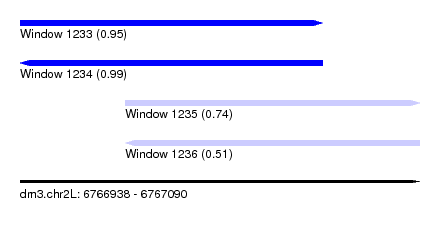
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,766,938 – 6,767,090 |
| Length | 152 |
| Max. P | 0.993045 |

| Location | 6,766,938 – 6,767,053 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Shannon entropy | 0.46868 |
| G+C content | 0.50434 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -19.78 |
| Energy contribution | -19.36 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
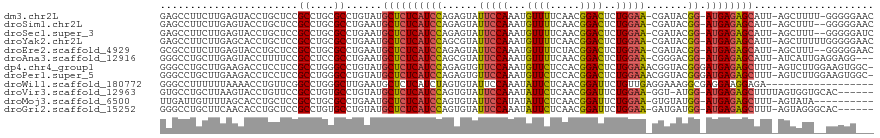
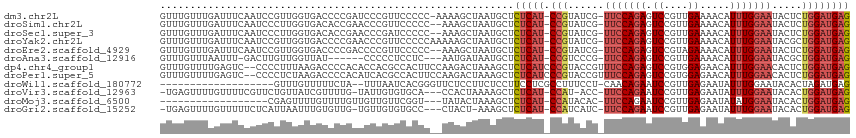
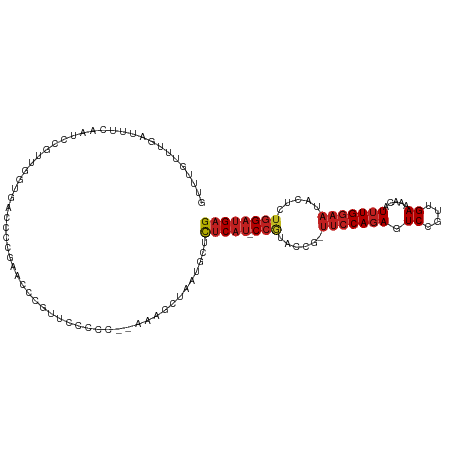
>dm3.chr2L 6766938 115 + 23011544 GAGCCUUCUUGAGUACCUGCUCCGCCUGCGCCUGUAUGCUCUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUU-AGCUUUU-GGGGGAAC ...(((((..((((....((.......))......((((((((((((......(((((...((((.....))))..)))))-......))-)))))))))).-.))))..-)))))... ( -37.20, z-score = -1.82, R) >droSim1.chr2L 6567418 114 + 22036055 GAGCCUUCUUGAGUACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUU-AGCUUU--GGGGGAAC ....................(((.((...((...(((((((((((((......(((((...((((.....))))..)))))-......))-)))))))))))-.))...--)).))).. ( -37.80, z-score = -2.01, R) >droSec1.super_3 2305596 114 + 7220098 GAGCCUUCUUGAGUACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUU-AGCUUU--GGGGGAUC ....................(((.((...((...(((((((((((((......(((((...((((.....))))..)))))-......))-)))))))))))-.))...--)).))).. ( -37.60, z-score = -1.72, R) >droYak2.chr2L 16181809 116 - 22324452 GAGCCUUCUUGAGCACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGCGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUU-AGCUUUUUGGGGGAAC ...(((((..((((....((.......)).....(((((((((((((..(((..((((...((((.....))))..)))))-))....))-)))))))))))-.))))...)))))... ( -41.30, z-score = -2.85, R) >droEre2.scaffold_4929 15683137 114 + 26641161 GCGCCUUCUUGAGUACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGAGUAUUCCAAAUGUUUUCUACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUU-AGCUUU--GGGGGAAC ....................(((.((...((...(((((((((((((......(((((...((((.....))))..)))))-......))-)))))))))))-.))...--)).))).. ( -37.80, z-score = -2.01, R) >droAna3.scaffold_12916 8344923 113 + 16180835 GGGCCUGCUUGAGUACCUUUUCCGCCUCCGCCUGAAUGCUCUCAUCCAGCGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGGGACGG-AUGAGAGCAUU-AUCAUUGAGGAGG--- ((..((.....))..))....((.((((......(((((((((((((..(.(.(((((...((((.....))))..)))))-.).)..))-)))))))))))-......)))).))--- ( -40.60, z-score = -2.37, R) >dp4.chr4_group1 3871607 117 + 5278887 GGGCCUGCUUGAAGACCUCCUCCGCCUGGGCCUGUAUGCUCUCAUCCAGAGUGUUCCAAAUGUUCUCCACGGACUCUGGAAACGGUACGGGAUGAGAGCUUU-AGUCUUGGAAGUGGC- (((.((......)).)))...(((((..((.(((...((((((((((..(.(((((((...((((.....))))..))).)))).)...))))))))))..)-)).))..)..)))).- ( -42.20, z-score = -1.03, R) >droPer1.super_5 5414761 117 - 6813705 GGGCCUGCUUGAAGACCUCCUCCGCCUGGGCCUGUAUGCUCUCAUCCAGAGUGUUCCAAAUGUUCUCCACGGACUCUGGAAACGGUACGGGAUGAGAGCUUU-AGUCUUGGAAGUGGC- (((.((......)).)))...(((((..((.(((...((((((((((..(.(((((((...((((.....))))..))).)))).)...))))))))))..)-)).))..)..)))).- ( -42.20, z-score = -1.03, R) >droWil1.scaffold_180772 7389705 101 + 8906247 GGGCCUUUUUUAAAACCUGUUCGGCCUGGGCUUGAAUGCUCUCAUCUAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGUUGAGGAAAGGCGAGGAAGGAGA------------------ ...((((........((.....))(((.(.(((((((((.((.....)).))))))......((((((((((...))))))))))))).).)))))))...------------------ ( -28.10, z-score = -0.22, R) >droVir3.scaffold_12963 16321808 110 - 20206255 GUGCCUGCUUAAGUACCUGUUCCGCCUGUGCCUGUAUGCUCUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGGAA-GGU-AUGG-AUGAGAGCUUUUAGUGGUGCAC------ (((((..((...((((..(.....)..))))......(((((((((((.....(((((...((((.....))))..)))))-...-.)))-))))))))....))..).))))------ ( -38.70, z-score = -3.60, R) >droMoj3.scaffold_6500 24616700 106 - 32352404 UUGAUUGUUUUAGCACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGUGUAUUCCAUAUAUUCUCAACGGAUUCUGGAA-GUGUAUGG-AUGAGAGCUUU-AGUAUA---------- ............(((...((...)).)))..(((((.(((((((((((.....(((((...((((.....))))..)))))-.....)))-)))))))))))-))....---------- ( -33.30, z-score = -2.92, R) >droGri2.scaffold_15252 1509361 110 + 17193109 GGGCCUGCUUCAACACCUGCUCCGCCUGUGCCUGUAUGCUCUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGGAA-GAUGAUGG-AUGAGAGCUUU-AGUAGGGCAC------ ((((..((..........))...))))((((((....(((((((((((.....(((((...((((.....))))..)))))-.....)))-))))))))...-....))))))------ ( -43.50, z-score = -3.87, R) >consensus GGGCCUGCUUGAGUACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGAGUAUUCCAAAUGUUCUCAACGGACUCUGGAA_CGAUACGG_AUGAGAGCAUU_AGCUUU__GGGGG___ .......................((....))......((((((((((......(((((...((((.....))))..))))).......)).)))))))).................... (-19.78 = -19.36 + -0.42)
| Location | 6,766,938 – 6,767,053 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Shannon entropy | 0.46868 |
| G+C content | 0.50434 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6766938 115 - 23011544 GUUCCCCC-AAAAGCU-AAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAGAGCAUACAGGCGCAGGCGGAGCAGGUACUCAAGAAGGCUC (((((.((-....(((-.((((((((((-(((....(-(((((((.((....)).....))))))))....)))))))))))))...)))...)).))))).................. ( -41.70, z-score = -3.42, R) >droSim1.chr2L 6567418 114 - 22036055 GUUCCCCC--AAAGCU-AAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUACUCAAGAAGGCUC (((((.((--...(((-(((((((((((-(((....(-(((((((.((....)).....))))))))....))))))))))))))..)))...)).))))).................. ( -43.60, z-score = -3.78, R) >droSec1.super_3 2305596 114 - 7220098 GAUCCCCC--AAAGCU-AAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUACUCAAGAAGGCUC ..(((.((--...(((-(((((((((((-(((....(-(((((((.((....)).....))))))))....))))))))))))))..)))...)).))).................... ( -39.40, z-score = -2.47, R) >droYak2.chr2L 16181809 116 + 22324452 GUUCCCCCAAAAAGCU-AAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACGCUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUGCUCAAGAAGGCUC (((((.((.....(((-(((((((((((-(((....(-(((((((.((....)).....))))))))....))))))))))))))..)))...)).)))))....(((......))).. ( -43.80, z-score = -3.30, R) >droEre2.scaffold_4929 15683137 114 - 26641161 GUUCCCCC--AAAGCU-AAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUAGAAAACAUUUGGAAUACUCUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUACUCAAGAAGGCGC (((((.((--...(((-(((((((((((-(((....(-(((((((.((....)).....))))))))....))))))))))))))..)))...)).))))).................. ( -43.80, z-score = -3.90, R) >droAna3.scaffold_12916 8344923 113 - 16180835 ---CCUCCUCAAUGAU-AAUGCUCUCAU-CCGUCCCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACGCUGGAUGAGAGCAUUCAGGCGGAGGCGGAAAAGGUACUCAAGCAGGCCC ---((.((((..(((.-.((((((((((-(((.(..(-(((((((.((....)).....))))))))..).))))))))))))))))....)))).))....(((.((.....))))). ( -41.50, z-score = -2.88, R) >dp4.chr4_group1 3871607 117 - 5278887 -GCCACUUCCAAGACU-AAAGCUCUCAUCCCGUACCGUUUCCAGAGUCCGUGGAGAACAUUUGGAACACUCUGGAUGAGAGCAUACAGGCCCAGGCGGAGGAGGUCUUCAAGCAGGCCC -....(((((......-...((((((((((.(....(((((((((.((......))...))))))).)).).))))))))))......((....))))))).(((((......))))). ( -41.10, z-score = -1.77, R) >droPer1.super_5 5414761 117 + 6813705 -GCCACUUCCAAGACU-AAAGCUCUCAUCCCGUACCGUUUCCAGAGUCCGUGGAGAACAUUUGGAACACUCUGGAUGAGAGCAUACAGGCCCAGGCGGAGGAGGUCUUCAAGCAGGCCC -....(((((......-...((((((((((.(....(((((((((.((......))...))))))).)).).))))))))))......((....))))))).(((((......))))). ( -41.10, z-score = -1.77, R) >droWil1.scaffold_180772 7389705 101 - 8906247 ------------------UCUCCUUCCUCGCCUUUCCUCAACAGAAUCCGUUGAGAAUAUUUGGAAUACACUAGAUGAGAGCAUUCAAGCCCAGGCCGAACAGGUUUUAAAAAAGGCCC ------------------...........((((((.((((((.......))))))..(((((((......)))))))(((((.(((..((....)).)))...)))))...)))))).. ( -21.70, z-score = -0.53, R) >droVir3.scaffold_12963 16321808 110 + 20206255 ------GUGCACCACUAAAAGCUCUCAU-CCAU-ACC-UUCCAGAAUCCGUUGAGAAUAUUUGGAAUACACUGGAUGAGAGCAUACAGGCACAGGCGGAACAGGUACUUAAGCAGGCAC ------(((((((.......((((((((-(((.-...-(((((((.((......))...))))))).....)))))))))))......((....))......))).((.....)))))) ( -32.90, z-score = -2.91, R) >droMoj3.scaffold_6500 24616700 106 + 32352404 ----------UAUACU-AAAGCUCUCAU-CCAUACAC-UUCCAGAAUCCGUUGAGAAUAUAUGGAAUACACUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUGCUAAAACAAUCAA ----------......-...((((((((-(((.....-(((((.(.((......)).)...))))).....))))))))))).....(((((..((...))..)))))........... ( -32.80, z-score = -3.45, R) >droGri2.scaffold_15252 1509361 110 - 17193109 ------GUGCCCUACU-AAAGCUCUCAU-CCAUCAUC-UUCCAGAAUCCGUUGAGAAUAUUUGGAAUACACUGGAUGAGAGCAUACAGGCACAGGCGGAGCAGGUGUUGAAGCAGGCCC ------(((((.....-...((((((((-(((.....-(((((((.((......))...))))))).....))))))))))).....)))))....((.((..((......))..)))) ( -37.52, z-score = -2.38, R) >consensus ___CCCCC__AAAGCU_AAUGCUCUCAU_CCGUAUCG_UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUACUCAAGAAGGCCC ....................((((((((.(((......(((((((.((....)).....))))))).....)))))))))))......((....))....................... (-19.93 = -20.52 + 0.59)

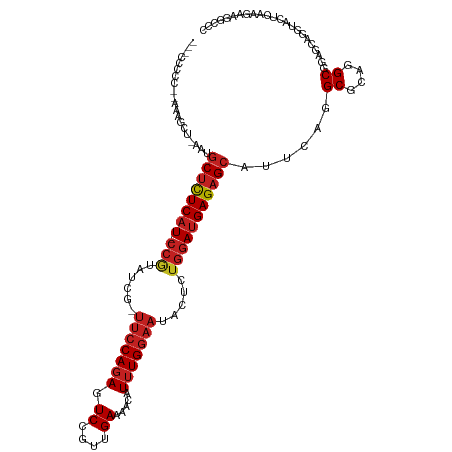
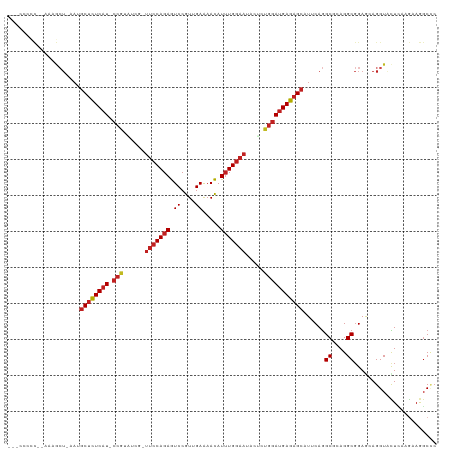
| Location | 6,766,978 – 6,767,090 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.68 |
| Shannon entropy | 0.74417 |
| G+C content | 0.44256 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -12.17 |
| Energy contribution | -11.50 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6766978 112 + 23011544 CUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUUAGCUUUU-GGGGGAACGGGAUCGGGGUCACCAACGGAUUGAAAUCAAACAAAC ....(((((((...(((...((....))..)))))))))).-((((.((.-..((((((....))))))-........(((((....))).))..)).))))............. ( -25.60, z-score = 0.00, R) >droSim1.chr2L 6567458 111 + 22036055 CUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUUAGCUUU--GGGGGAACGGGUUCGGUGUCACCAAGGGAUUGAAAUCAAACAAAC (((.(((((((...(((...((....))..)))))))))).-.(((((.(-((.(((((....)))))--.)(....)....)).))))).....))).(((....)))...... ( -26.90, z-score = -0.17, R) >droSec1.super_3 2305636 111 + 7220098 CUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUUAGCUUU--GGGGGAUCGGGUUCGGUGUCACCAAGGGAUUGAAAUCAAACAAAC (((((((......(((((...((((.....))))..)))))-......))-)))))........((((--((((.((((....)))).)).))))))..(((....)))...... ( -28.90, z-score = -0.63, R) >droYak2.chr2L 16181849 113 - 22324452 CUCAUCCAGCGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUUAGCUUUUUGGGGGAACGGGUUCGGGGUCACCAACGGAUUGAAAUCAAACAAAC (((((((..(((..((((...((((.....))))..)))))-))....))-)))))(((....))).((((...((...((((((((....))..))))))....))...)))). ( -25.80, z-score = 0.18, R) >droEre2.scaffold_4929 15683177 111 + 26641161 CUCAUCCAGAGUAUUCCAAAUGUUUUCUACGGACUCUGGAA-CGAUACGG-AUGAGAGCAUUAGCUUU--GGGGGAACGGGGUCGGGGUCACCAACGGAUUGAAAUCAAACAAAC .((((((((((...(((.............)))))))))).-((((.((.-.(.(((((....)))))--.).....))..))))((....)).......)))............ ( -24.42, z-score = 0.68, R) >droAna3.scaffold_12916 8344963 103 + 16180835 CUCAUCCAGCGUAUUCCAAAUGUUUUCAACGGACUCUGGAA-CGGGACGG-AUGAGAGCAUUAUCAUU---GAGGAGGGGG------AUAACCAACAAGUC-AAAUUAAACAAAC (((((((..(.(.(((((...((((.....))))..)))))-.).)..))-)))))..........((---((.(..((..------....))..)...))-))........... ( -22.20, z-score = -0.63, R) >dp4.chr4_group1 3871647 113 + 5278887 CUCAUCCAGAGUGUUCCAAAUGUUCUCCACGGACUCUGGAAACGGUACGGGAUGAGAGCUUUAGUCUUGGAAGUGGCGUGGUGUGGGGUCUUAAAGGGG--GACUCAAAACAAAC (((((((..(.(((((((...((((.....))))..))).)))).)...))))))).(((...(((........)))..)))((.(((((((.....))--)))))...)).... ( -33.90, z-score = -0.55, R) >droPer1.super_5 5414801 113 - 6813705 CUCAUCCAGAGUGUUCCAAAUGUUCUCCACGGACUCUGGAAACGGUACGGGAUGAGAGCUUUAGUCUUGGAAGUGGCGUGAUGUGGGGUCUUAGAGGGG--GACUCAAAACAAAC (((((((..(.(((((((...((((.....))))..))).)))).)...))))))).......(((........)))....(((.(((((((.....))--)))))...)))... ( -32.90, z-score = -0.08, R) >droWil1.scaffold_180772 7389745 93 + 8906247 CUCAUCUAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGUUG-AGGAAAGGCGAGGAAGGAGAAGGAGAACCCGUGAUUAAA--UAGAAAAACAAAC------------------- ....((((...((.((......((((((((((...))))))-))))...(((.((..............))))))).))..--)))).........------------------- ( -17.04, z-score = 0.06, R) >droVir3.scaffold_12963 16321848 107 - 20206255 CUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGGAA-GGU-AUGG-AUGAGAGCUUUUAGUGG---UGCACACAAUA-CAAAACGAUAACAGAACGAAAACAAAACUCA- ((((((((.....(((((...((((.....))))..)))))-...-.)))-)))))........(((.---....)))....-...............................- ( -22.90, z-score = -1.39, R) >droMoj3.scaffold_6500 24616740 92 - 32352404 CUCAUCCAGUGUAUUCCAUAUAUUCUCAACGGAUUCUGGAA-GUGUAUGG-AUGAGAGCUUUAGUAUA---ACCGAACAACAACAAAACAAAACUCG------------------ ((((((((.....(((((...((((.....))))..)))))-.....)))-)))))............---..........................------------------ ( -20.30, z-score = -2.06, R) >droGri2.scaffold_15252 1509401 107 + 17193109 CUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGGAA-GAUGAUGG-AUGAGAGCUUU-AGUAG---GGCACACAACA-CAACACAAAUUAAUGAGAAAAACAAAACUCA- ((((((((.....(((((...((((.....))))..)))))-.....)))-))))).((((.-....)---)))........-.............((((..........))))- ( -24.20, z-score = -2.16, R) >consensus CUCAUCCAGAGUAUUCCAAAUGUUCUCAACGGACUCUGGAA_CGAUACGG_AUGAGAGCAUUAGCUUU__GGGGGAACGGGUUCGGGGUCACCAACGGAUUGAAAUCAAACAAAC (((((((......(((((...((((.....))))..))))).......)).)))))........................................................... (-12.17 = -11.50 + -0.66)

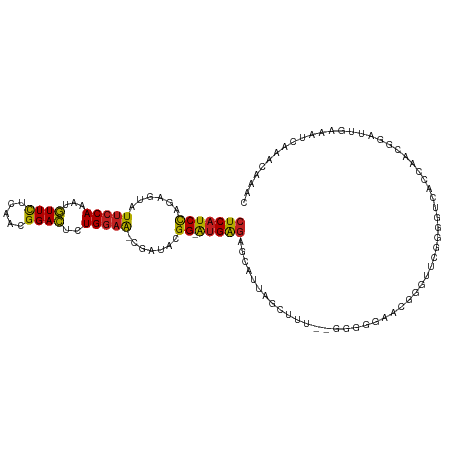
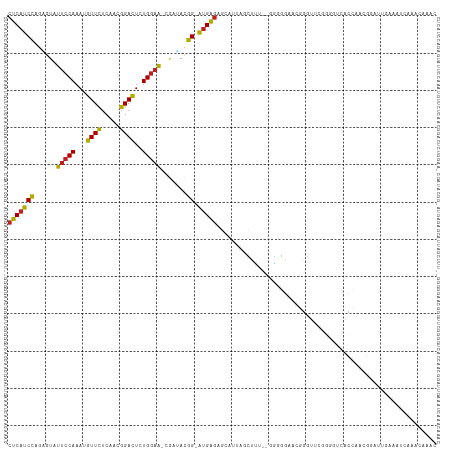
| Location | 6,766,978 – 6,767,090 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.68 |
| Shannon entropy | 0.74417 |
| G+C content | 0.44256 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -9.60 |
| Energy contribution | -10.11 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6766978 112 - 23011544 GUUUGUUUGAUUUCAAUCCGUUGGUGACCCCGAUCCCGUUCCCCC-AAAAGCUAAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAG .....((((..........(((((.....)))))..........)-)))(((....)))(((((-(((....(-(((((((.((....)).....))))))))....)))))))) ( -22.65, z-score = -0.59, R) >droSim1.chr2L 6567458 111 - 22036055 GUUUGUUUGAUUUCAAUCCCUUGGUGACACCGAACCCGUUCCCCC--AAAGCUAAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAG (((((.(((....))).....((....)).)))))..........--..(((....)))(((((-(((....(-(((((((.((....)).....))))))))....)))))))) ( -24.20, z-score = -1.08, R) >droSec1.super_3 2305636 111 - 7220098 GUUUGUUUGAUUUCAAUCCCUUGGUGACACCGAACCCGAUCCCCC--AAAGCUAAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAG (((((.(((....))).....((....)).)))))..........--..(((....)))(((((-(((....(-(((((((.((....)).....))))))))....)))))))) ( -24.20, z-score = -0.97, R) >droYak2.chr2L 16181849 113 + 22324452 GUUUGUUUGAUUUCAAUCCGUUGGUGACCCCGAACCCGUUCCCCCAAAAAGCUAAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACGCUGGAUGAG ..................(((((((......(((....))).........)))))))..(((((-(((....(-(((((((.((....)).....))))))))....)))))))) ( -22.76, z-score = -0.40, R) >droEre2.scaffold_4929 15683177 111 - 26641161 GUUUGUUUGAUUUCAAUCCGUUGGUGACCCCGACCCCGUUCCCCC--AAAGCUAAUGCUCUCAU-CCGUAUCG-UUCCAGAGUCCGUAGAAAACAUUUGGAAUACUCUGGAUGAG ...................(((((.....)))))...........--..(((....)))(((((-(((....(-(((((((.((....)).....))))))))....)))))))) ( -24.30, z-score = -1.23, R) >droAna3.scaffold_12916 8344963 103 - 16180835 GUUUGUUUAAUUU-GACUUGUUGGUUAU------CCCCCUCCUC---AAUGAUAAUGCUCUCAU-CCGUCCCG-UUCCAGAGUCCGUUGAAAACAUUUGGAAUACGCUGGAUGAG (((((((....((-((......((....------))......))---)).))))).)).(((((-(((.(..(-(((((((.((....)).....))))))))..).)))))))) ( -21.00, z-score = -0.92, R) >dp4.chr4_group1 3871647 113 - 5278887 GUUUGUUUUGAGUC--CCCCUUUAAGACCCCACACCACGCCACUUCCAAGACUAAAGCUCUCAUCCCGUACCGUUUCCAGAGUCCGUGGAGAACAUUUGGAACACUCUGGAUGAG ((..((((((((..--....))))))))...))..........................(((((((.(....(((((((((.((......))...))))))).)).).))))))) ( -20.20, z-score = 0.71, R) >droPer1.super_5 5414801 113 + 6813705 GUUUGUUUUGAGUC--CCCCUCUAAGACCCCACAUCACGCCACUUCCAAGACUAAAGCUCUCAUCCCGUACCGUUUCCAGAGUCCGUGGAGAACAUUUGGAACACUCUGGAUGAG ((..((((((((..--...))).)))))...))..........................(((((((.(....(((((((((.((......))...))))))).)).).))))))) ( -20.60, z-score = 0.82, R) >droWil1.scaffold_180772 7389745 93 - 8906247 -------------------GUUUGUUUUUCUA--UUUAAUCACGGGUUCUCCUUCUCCUUCCUCGCCUUUCCU-CAACAGAAUCCGUUGAGAAUAUUUGGAAUACACUAGAUGAG -------------------...(((..(((((--..((.(((((((((((.......................-....)))))))).)))...))..))))).)))......... ( -15.33, z-score = -0.16, R) >droVir3.scaffold_12963 16321848 107 + 20206255 -UGAGUUUUGUUUUCGUUCUGUUAUCGUUUUG-UAUUGUGUGCA---CCACUAAAAGCUCUCAU-CCAU-ACC-UUCCAGAAUCCGUUGAGAAUAUUUGGAAUACACUGGAUGAG -.(((((((......((..(((...((.....-...))...)))---..))..)))))))((((-(((.-...-(((((((.((......))...))))))).....))))))). ( -20.60, z-score = -0.28, R) >droMoj3.scaffold_6500 24616740 92 + 32352404 ------------------CGAGUUUUGUUUUGUUGUUGUUCGGU---UAUACUAAAGCUCUCAU-CCAUACAC-UUCCAGAAUCCGUUGAGAAUAUAUGGAAUACACUGGAUGAG ------------------.(((((((((...(..(.....)..)---...)).)))))))((((-(((.....-(((((.(.((......)).)...))))).....))))))). ( -19.90, z-score = -0.79, R) >droGri2.scaffold_15252 1509401 107 - 17193109 -UGAGUUUUGUUUUUCUCAUUAAUUUGUGUUG-UGUUGUGUGCC---CUACU-AAAGCUCUCAU-CCAUCAUC-UUCCAGAAUCCGUUGAGAAUAUUUGGAAUACACUGGAUGAG -.(((((((((....(.(((..((.......)-)...))).)..---..)).-)))))))((((-(((.....-(((((((.((......))...))))))).....))))))). ( -21.40, z-score = -0.59, R) >consensus GUUUGUUUGAUUUCAAUCCGUUGGUGACCCCGAACCCGUUCCCCC__AAAGCUAAUGCUCUCAU_CCGUACCG_UUCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAG ...........................................................(((((.(((......(((((((.((....)).....))))))).....)))))))) ( -9.60 = -10.11 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:54 2011