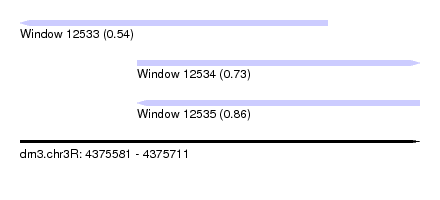
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,375,581 – 4,375,711 |
| Length | 130 |
| Max. P | 0.864906 |

| Location | 4,375,581 – 4,375,681 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.38 |
| Shannon entropy | 0.55348 |
| G+C content | 0.51752 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -13.63 |
| Energy contribution | -12.39 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4375581 100 - 27905053 ----------GCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUG--UGGCAGGGCACACAUGUUAGGGGUAUCCCCUCUUCCCCACC-ACGUGGCCAGCU--UAAUAAACCUA ----------(((((((((........)))))))((((((.....)--))))).(((.(((.((...((((...........))))..)-).)))))).)).--........... ( -35.10, z-score = -1.54, R) >droWil1.scaffold_181089 936797 114 - 12369635 UUUCAAGUCAGUCAUUGUCCUUUGGUUGGUUGUUUUUUGGGUUUGGGUUGGCCUGGC-UGGGUGCCAAAUAUUUUGACUUGUCUAAACCACAGUAUACAGUACUUAUUUAACUUA ...(((((((((((....(((..(.(..(.......)..).)..))).)))).((((-.....)))).......)))))))..........(((((...)))))........... ( -22.00, z-score = 1.27, R) >droPer1.super_3 6120687 95 - 7375914 ----------GCCAUUGCCCUC-UGCUGGCAAUGUGCCACUUUCCU--UGGCAGCACACACAUGCUUGGGGUAUCCGUACCCUCG-----GCUCCAAAAACA--UAAUAAACUUA ----------(((((((((...-....)))))))(((((.......--)))))))........(((.((((((....)))))).)-----))..........--........... ( -28.10, z-score = -2.04, R) >dp4.chr2 23348373 95 - 30794189 ----------GCCAUUGCCCUC-UGCUGGCAAUGUGCCACUUUCCU--UGGCAGCACACACAUGCUUGGGGUAUCCGUACCCUCG-----GCUCCAAAAACA--UAAUAAACUUC ----------(((((((((...-....)))))))(((((.......--)))))))........(((.((((((....)))))).)-----))..........--........... ( -28.10, z-score = -2.29, R) >droAna3.scaffold_13340 19681344 100 + 23697760 ----------GCCAUUGCCCUC-UGCUGGCAGUGUGCCACAUUCUG--UGGCACUGCACACAUGUUCGGGGUGUCCUCUCCCCCUUUCUAAUGUGGCCAACA--UAAUAAACUUA ----------...((((((...-....))))))(((((((.....)--)))))).((.(((((....((((.(......)))))......))))))).....--........... ( -32.50, z-score = -2.33, R) >droEre2.scaffold_4770 547062 99 + 17746568 ----------GCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUG--UGGCAGGGCACACAUGCUUGGGGUA-CCCUUUCUCCCCUCC-ACGUGGCCAGCA--UAAUAAACCUA ----------(((((((((........)))))))((((((.....)--))))).(((.(((.((...((((..-........))))..)-).)))))).)).--........... ( -36.40, z-score = -1.90, R) >droYak2.chr3R 8448775 99 - 28832112 ----------GCCAUUGCCCUCUUGCUGGCAAUGUGCCACAUUCUG--UGGCAGGGCACACAUGCUAGGGGUAUCCUCUUUUCCCU-CC-ACGUGGCUAGCA--UAAUAAACCUA ----------(((((((((........)))))).((((((.....)--))))).)))....((((((((((...........))).-((-....))))))))--).......... ( -34.00, z-score = -1.58, R) >droSec1.super_0 3656265 100 + 21120651 ----------GCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUG--UGGCAGGGCACACAUGCUAGGGGUGUUCCCCAUUCCCCACC-ACGUGGCCAGCG--UAAUAAACCUA ----------(((((((((........)))))).((((((.....)--))))).)))....(((((.((((....)))).....((((.-..))))..))))--).......... ( -36.80, z-score = -1.12, R) >droSim1.chr3R 10618878 100 + 27517382 ----------GCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUG--UGGCAGGGCACACAUGCUAGGGGUAUCCCCCAUUCCCCACC-ACGUGGCCAGCG--UAAUAAACCUA ----------(((((((((........)))))).((((((.....)--))))).)))....(((((.((((....)))).....((((.-..))))..))))--).......... ( -36.30, z-score = -1.25, R) >consensus __________GCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUG__UGGCAGGGCACACAUGCUAGGGGUAUCCCCUCCUCCCCACC_ACGUGGCCAGCA__UAAUAAACCUA ..........(((((((((........)))))))(((((.........)))))..........))..((((........))))................................ (-13.63 = -12.39 + -1.24)

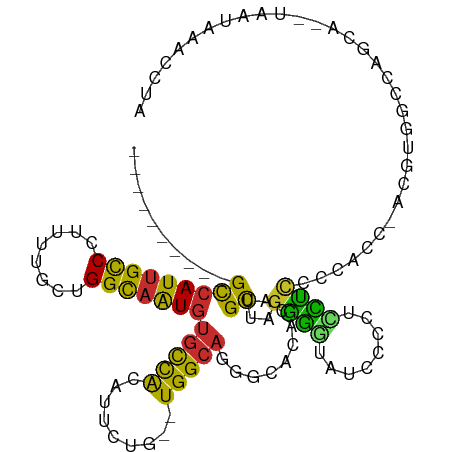
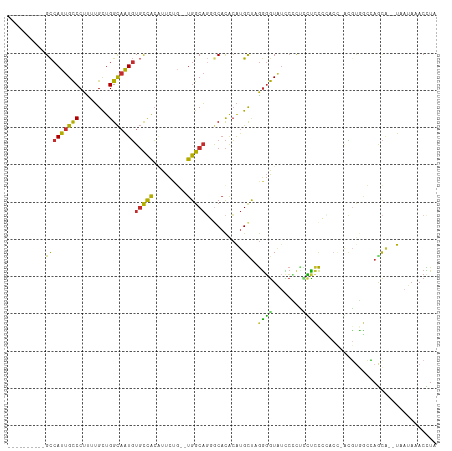
| Location | 4,375,619 – 4,375,711 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.85 |
| Shannon entropy | 0.31983 |
| G+C content | 0.49072 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -21.52 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4375619 92 + 27905053 AUACCCCUAACAUGUGUGCCCUGCCACAGAAUGUGGCACAUUGCCAGCAAAAGGGCAAUGGCAGCUA-GUCGAAAAAAUAAUAGAGGUAUGUA--- (((((.(((.......(((((((((((.....)))))...(((....))).)))))).((((.....-)))).........))).)))))...--- ( -29.00, z-score = -1.84, R) >dp4.chr2 23348407 92 + 30794189 AUACCCCAAGCAUGUGUGUGCUGCCAAGGAAAGUGGCACAUUGCCAGCA-GAGGGCAAUGGCAGCGGCAACGGACAAAAAAUAAAGGAAAAUA--- ............(((.((((((((((.......))((.(((((((....-...))))))))).))))).))).))).................--- ( -25.00, z-score = -0.16, R) >droAna3.scaffold_13340 19681383 94 - 23697760 ACACCCCGAACAUGUGUGCAGUGCCACAGAAUGUGGCACACUGCCAGCA-GAGGGCAAUGGCAGCUC-GGCGAAAAAAUAAUAGAAAUGUUAGGAG ...((((((...........(((((((.....))))))).((((((((.-....))..)))))).))-))........(((((....))))))).. ( -31.80, z-score = -2.40, R) >droEre2.scaffold_4770 547099 88 - 17746568 GUACCCCAAGCAUGUGUGCCCUGCCACAGAAUGUGGCACAUUGCCAGCAAAAGGGCAAUGGCAGCUA-GUCGAAAAAAUAAUAGAGGAC------- .....((.(((.(((.(((((((((((.....)))))...(((....))).))))))...)))))).-.((............))))..------- ( -25.70, z-score = -1.22, R) >droYak2.chr3R 8448812 86 + 28832112 AUACCCCUAGCAUGUGUGCCCUGCCACAGAAUGUGGCACAUUGCCAGCAAGAGGGCAAUGGCAGCUA-GUCGAAAA--UAAUAGAGGUA------- .((((.(((((.(((.(((((((((((.....)))))...(((....))).))))))...)))))))-)((.....--.....))))))------- ( -30.80, z-score = -2.24, R) >droSec1.super_0 3656303 94 - 21120651 ACACCCCUAGCAUGUGUGCCCUGCCACAGAAUGUGGCACAUUGCCAGCAAAAGGGCAAUGGCAGCUG-GUCGAAAAAAUAAUAGAGGUGUAUGCA- (((((.(((((.(((.(((((((((((.....)))))...(((....))).))))))...)))))))-)((............))))))).....- ( -33.60, z-score = -1.57, R) >droSim1.chr3R 10618916 94 - 27517382 AUACCCCUAGCAUGUGUGCCCUGCCACAGAAUGUGGCACAUUGCCAGCAAAAGGGCAAUGGCAGCUA-GUCGAAAAAAUAAUAGAGGUGUAUGCA- (((((.(((((.(((.(((((((((((.....)))))...(((....))).))))))...)))))))-)((............))))))).....- ( -32.00, z-score = -1.76, R) >consensus AUACCCCUAGCAUGUGUGCCCUGCCACAGAAUGUGGCACAUUGCCAGCAAAAGGGCAAUGGCAGCUA_GUCGAAAAAAUAAUAGAGGUAUAUG___ .......((((.(((......((((((.....))))))(((((((........))))))))))))))............................. (-21.52 = -22.30 + 0.78)

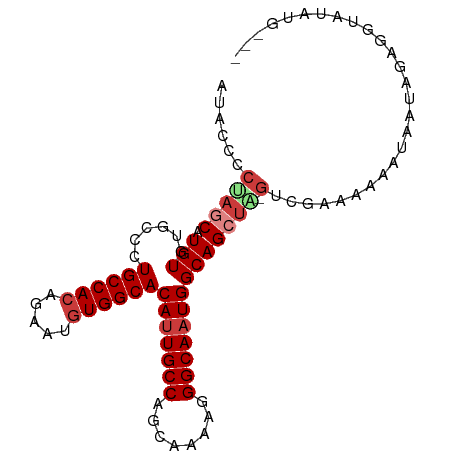

| Location | 4,375,619 – 4,375,711 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.85 |
| Shannon entropy | 0.31983 |
| G+C content | 0.49072 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.91 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4375619 92 - 27905053 ---UACAUACCUCUAUUAUUUUUUCGAC-UAGCUGCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUGUGGCAGGGCACACAUGUUAGGGGUAU ---...(((((((((.............-..(((..(((((((........)))))))((((((.....)))))).)))((....))))))))))) ( -30.90, z-score = -2.39, R) >dp4.chr2 23348407 92 - 30794189 ---UAUUUUCCUUUAUUUUUUGUCCGUUGCCGCUGCCAUUGCCCUC-UGCUGGCAAUGUGCCACUUUCCUUGGCAGCACACACAUGCUUGGGGUAU ---.....((((........(((..(((((((..(((((((((...-....))))))).)).........)))))))..))).......))))... ( -22.66, z-score = -0.04, R) >droAna3.scaffold_13340 19681383 94 + 23697760 CUCCUAACAUUUCUAUUAUUUUUUCGCC-GAGCUGCCAUUGCCCUC-UGCUGGCAGUGUGCCACAUUCUGUGGCACUGCACACAUGUUCGGGGUGU ((((.(((((..................-..(((((((..((....-.)))))))))(((((((.....))))))).......))))).))))... ( -32.00, z-score = -2.59, R) >droEre2.scaffold_4770 547099 88 + 17746568 -------GUCCUCUAUUAUUUUUUCGAC-UAGCUGCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUGUGGCAGGGCACACAUGCUUGGGGUAC -------((((((...............-.......(((((((........)))))))((((((.....))))))(((((....))))))))).)) ( -25.80, z-score = -0.73, R) >droYak2.chr3R 8448812 86 - 28832112 -------UACCUCUAUUA--UUUUCGAC-UAGCUGCCAUUGCCCUCUUGCUGGCAAUGUGCCACAUUCUGUGGCAGGGCACACAUGCUAGGGGUAU -------(((((......--.......(-((((((((((((((........)))))).((((((.....)))))).)))).....)))))))))). ( -29.91, z-score = -1.83, R) >droSec1.super_0 3656303 94 + 21120651 -UGCAUACACCUCUAUUAUUUUUUCGAC-CAGCUGCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUGUGGCAGGGCACACAUGCUAGGGGUGU -.....(((((((((............(-(......(((((((........)))))))((((((.....))))))))(((....)))))))))))) ( -34.50, z-score = -2.09, R) >droSim1.chr3R 10618916 94 + 27517382 -UGCAUACACCUCUAUUAUUUUUUCGAC-UAGCUGCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUGUGGCAGGGCACACAUGCUAGGGGUAU -...((((.((((............))(-((((((((((((((........)))))).((((((.....)))))).)))).....))))))))))) ( -29.40, z-score = -1.07, R) >consensus ___CAUACACCUCUAUUAUUUUUUCGAC_UAGCUGCCAUUGCCCUUUUGCUGGCAAUGUGCCACAUUCUGUGGCAGGGCACACAUGCUAGGGGUAU .......((((((.......................(((((((........)))))))((((((.....)))))).((((....)))).)))))). (-23.66 = -23.91 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:42 2011