
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,374,509 – 4,374,669 |
| Length | 160 |
| Max. P | 0.729891 |

| Location | 4,374,509 – 4,374,604 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.45 |
| Shannon entropy | 0.23468 |
| G+C content | 0.41736 |
| Mean single sequence MFE | -16.33 |
| Consensus MFE | -13.77 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
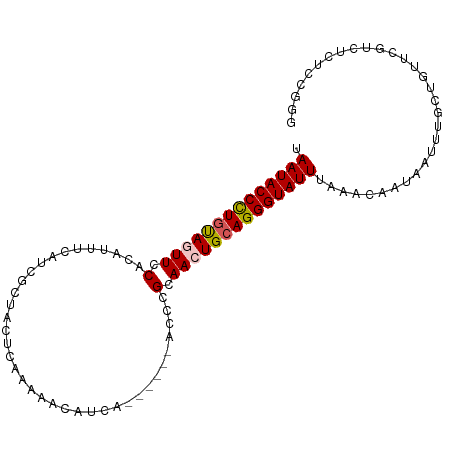
>dm3.chr3R 4374509 95 + 27905053 UAAUACCCUAUAGUUCCACAUUUCAUCGCUACUCAAAAACAUCA------ACCCGCAACUGCAGGGUAUUUAAACAAUAAUUUGCUGUUCGUCUCUCCGGG .((((((((.(((((.(...........................------....).))))).))))))))............................... ( -11.18, z-score = 0.04, R) >droEre2.scaffold_4770 546001 88 - 17746568 UAAUACCUUGUAGUUCCACAUUUCAUCGCUAC--AAAAUC-----------CCCGCAAGUGCAGGGUAUUUAAAUAUUAAUUUGCUGCUCGUCUCUCCGGG .......(((((((.............)))))--))....-----------((((..((.((((.(.(((........))).).))))....))...)))) ( -15.42, z-score = -0.88, R) >droYak2.chr3R 8447724 99 + 28832112 UAAUACCUUGCAGUUCCACAUUUCAUCGGUACUCGAAAGCAUCACAAAAUACCCGCAAGUGCAGGGUAUUUAAACAAUAAUUUGCUGCUCGUCUCCAGG-- .........(((((.............(((...(....).)))...((((((((((....)).))))))))............)))))...........-- ( -19.30, z-score = -1.34, R) >droSec1.super_0 3655201 95 - 21120651 UAAUACCCUGUAGUUCCACAUUUCAUCCCUACUCAAAAACAUCA------ACCCGCAACUGCAGGGUAUUUAAACAAUAAUUUGCUGUUCGUCUCUCCGGG .((((((((((((((.(...........................------....).))))))))))))))............................... ( -17.48, z-score = -2.33, R) >droSim1.chr3R 10617814 95 - 27517382 UAAUACCCUGUAGUUCCACAUAUCAUCGCUACUCAAAAACAUCA------ACCCGCAACUGCAGGGUAUUUAAACAAUAAUUUGCUGUUCGUCUCUCAGGG .((((((((((((((.(...........................------....).))))))))))))))..............(((.........))).. ( -18.28, z-score = -2.39, R) >consensus UAAUACCCUGUAGUUCCACAUUUCAUCGCUACUCAAAAACAUCA______ACCCGCAACUGCAGGGUAUUUAAACAAUAAUUUGCUGUUCGUCUCUCCGGG .((((((((((((((.(.....................................).))))))))))))))............................... (-13.77 = -13.97 + 0.20)


| Location | 4,374,572 – 4,374,669 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.19 |
| Shannon entropy | 0.49397 |
| G+C content | 0.40181 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.95 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.729891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
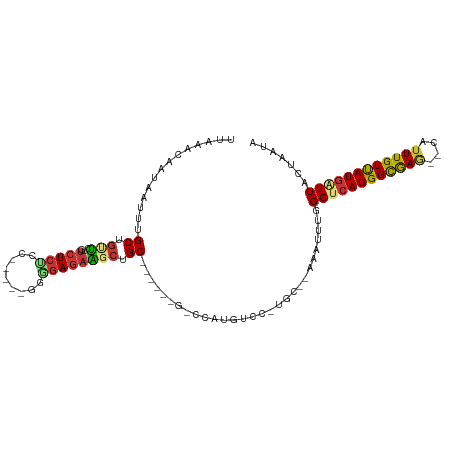
>dm3.chr3R 4374572 97 + 27905053 UUAAACAAUAAUUUGCUGUUC-GUCUCUCC-----GGGGAGAAGCUGC----AUGUCCAUGUCC-UGC--AAAUUUGGGUCAUGGCGAG--CAUUUGUUAUGAUUACUAAUA .......(((((....(((((-(..((((.-----...))))..)...----..(.(((((.((-(..--......))).)))))))))--))...)))))........... ( -24.80, z-score = -1.01, R) >dp4.chr2 23347310 112 + 30794189 UUAAACAAUCAUUUGCUGUUCCGUCUCUCCUCCACAGAGAGAGGCUGCCAUGGAGGACAUGUCUGUGCUGAAAUUUGGGUCAUGGCGAGAGCAUUUGUUAUGAUUAUUAUUA ......((((((..(((((((((((((((((....)).))))))).(((((((....((..((......))....))..)))))))).)))))...)).))))))....... ( -31.50, z-score = -0.85, R) >droAna3.scaffold_13340 19680231 103 - 23697760 UUAAACAAUAAUUUGCUGUUC-GUCUCUCC-----AGUGAGAGCCUGCACUGGAGAGCAUGUCCGUGC-AGAAUUUAGGUCAUGGUGAG--CAUUUGUUAUAGUUAUUAUUA ...(((.(((((....(((((-(.((((((-----((((........))))))))))((((.((....-........)).)))).))))--))...))))).)))....... ( -31.80, z-score = -2.81, R) >droEre2.scaffold_4770 546057 94 - 17746568 UUAAAUAUUAAUUUGCUGCUC-GUCUCUCC-----GGGGAGAAGCUGC----------AUGUCCAUGGAAGAAUUUGGGUCAUGGCGAG--CAUUUGUUAUGAUUACUAAUA ........(((((.(((((((-((.((((.-----...)))).....(----------(((((((..........)))).)))))))))--))...))...)))))...... ( -22.80, z-score = -0.43, R) >droYak2.chr3R 8447793 92 + 28832112 UUAAACAAUAAUUUGCUGCUC-GUCUCC-------AGGGAGAAGCUGC----------AUGUCCAUGGACGAAUUUGGGUCAUGGCGAG--CAUUUGUUAUGAUUACUAAUA .....(((.....(((.(((.-.((((.-------...))))))).))----------)((((....))))...)))(((((((((((.--...)))))))))))....... ( -24.70, z-score = -1.31, R) >droSec1.super_0 3655264 97 - 21120651 UUAAACAAUAAUUUGCUGUUC-GUCUCUCC-----GGGGAGAAGCUGC----AUGUCCAUGUCC-UGC--AAAUUUGGGUCAUGGCGAUA--AUUUGUUAUGCUUACUAAUA ...(((((.....(((.((..-.(((((..-----..))))).)).))----)((.(((((.((-(..--......))).)))))))...--..)))))............. ( -23.70, z-score = -1.16, R) >droSim1.chr3R 10617877 97 - 27517382 UUAAACAAUAAUUUGCUGUUC-GUCUCUCA-----GGGGAGAAGCUGC----AUAGCCAUGUCC-UGC--AAAUUUGGGUCAUGGCGAG--CAUUUGUUAUGCUUACUAAUA .............(((.((..-.(((((..-----..))))).)).))----)..((((((.((-(..--......))).))))))(((--(((.....))))))....... ( -30.80, z-score = -2.63, R) >droWil1.scaffold_181089 11434979 96 - 12369635 UUAAACAAUAAUUUGCUUCAUGCUUUUGUC----CUGGCAUCUUUAGCC-------CAAUUUUCUUGU---UAAGUGGGUCAUGGCAAA--CAUUUGUCAUGAUUAUUUCGA ...(((((.((((.(((..(((((......----..)))))....))).-------.))))...))))---).....(((((((((((.--...)))))))))))....... ( -20.70, z-score = -1.48, R) >consensus UUAAACAAUAAUUUGCUGUUC_GUCUCUCC_____GGGGAGAAGCUGC______G_CCAUGUCC_UGC__AAAUUUGGGUCAUGGCGAG__CAUUUGUUAUGAUUACUAAUA ..............((.(((...((((((.......))))))))).)).............................((((((((((((....))))))))))))....... (-13.57 = -13.95 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:40 2011