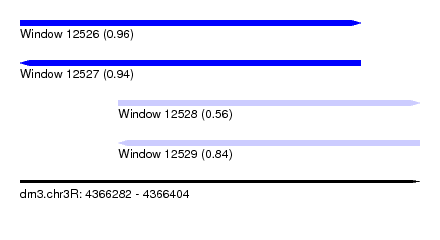
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,366,282 – 4,366,404 |
| Length | 122 |
| Max. P | 0.960839 |

| Location | 4,366,282 – 4,366,386 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.87 |
| Shannon entropy | 0.64756 |
| G+C content | 0.41247 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.50 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4366282 104 + 27905053 UUGCAACCACUGCCUUUGACCAA-UCGCCGAAAAAUGCUCGGCACCGCAGGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGCUUCUUUU---AUUUGAUUCAAAG ..(((.....)))((((((...(-(((((((.......))))).....(((((.((((((...(((((.....))))))))))).)))))..---....))))))))) ( -29.40, z-score = -3.26, R) >droSim1.chr3R 10609528 104 - 27517382 UUGCAACCACUGCCUUUGACCAA-UCGCCGAAAAAUGCUCGGCACCGCAGGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGCUUCUUUU---AUUAGAUUCAAAG ..(((.....)))((((((...(-(((((((.......))))).....(((((.((((((...(((((.....))))))))))).)))))..---....))))))))) ( -29.60, z-score = -3.40, R) >droSec1.super_0 3646920 104 - 21120651 UUGCAACCACUGCCUUUGACCAA-UCGCCGAAAAAUGCUCGGCACCGCAGGAAGCCGCAGAAUCAUAAAUAUUUUAUGCUGCGGCUUCUUUU---AUUAGAUUCAAAG ..(((.....)))((((((...(-(((((((.......))))).....((((((((((((...(((((.....)))))))))))))))))..---....))))))))) ( -36.20, z-score = -4.89, R) >droYak2.chr3R 8439341 104 + 28832112 UGGCAACCACUGCCUUUGACCAA-UCGCCGAACAAUGCUCGGCACCGCAGGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGCUUCUUUU---AUUUGAUUCAAAG .((((.....))))(((((...(-(((((((.......))))).....(((((.((((((...(((((.....))))))))))).)))))..---....)))))))). ( -31.10, z-score = -3.41, R) >droEre2.scaffold_4770 536724 104 - 17746568 UUGCAACCACUGCCUUUGACCAA-UCGCCGAAAAAUGCUCGGCACCGCAGGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGCUUCUUUU---AUUUGAUUCAAAG ..(((.....)))((((((...(-(((((((.......))))).....(((((.((((((...(((((.....))))))))))).)))))..---....))))))))) ( -29.40, z-score = -3.26, R) >droAna3.scaffold_13340 19661249 104 - 23697760 CCACUGCCACUGCCUUUGACCAA-UCGCCGAAAAAUGUGCUGCACCGCUCAAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGCUUUCUUU---AUUUGAUUCCACG ....................(((-.....((((..((.((......)).))...((((((...(((((.....))))))))))).))))...---..)))........ ( -13.40, z-score = 0.93, R) >dp4.chr2 23335962 93 + 30794189 ----------UGCCUUUGACCAA-UCGCCGAAAAAUAUUUUGCACCGCACCAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGCUUCCUUUU--AUUUGAUAUGG-- ----------.........((((-((((.(((.....))).))...........((((((...(((((.....))))))))))).........--....))).)))-- ( -13.90, z-score = 0.08, R) >droPer1.super_3 6108366 93 + 7375914 ----------UGCCUUUGACCAA-UCGCCGAAAAAUAUUUUGCACCGCACCAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGCUUCCUUUU--AUUUGAUAUGG-- ----------.........((((-((((.(((.....))).))...........((((((...(((((.....))))))))))).........--....))).)))-- ( -13.90, z-score = 0.08, R) >droWil1.scaffold_181089 926722 100 + 12369635 CAUCCAUCUAUCCUUUUGACCGU-UUUAUGUAUUUUGUAAACUUUGUUUGGCUACCGCAAAAAUAUAAAUAUUUUAGAUUGUGGCUUGUUUUUC-GCUGAGU------ .......................-................((((.((..(((..(((((((((((....)))))....))))))...)))....-)).))))------ ( -9.00, z-score = 1.88, R) >droMoj3.scaffold_6540 3478432 97 + 34148556 UUUCAACU-UUUUGGCUAGCCAAGUUGCCGCAAA---CCCAACGCC------AGCCGCAAUAUCAUAAAUAUUUUAUGUUGCGGCAUGUUUUGUAAUUCGUUUGUGU- ...(((((-...(((....)))))))).((((((---(.(((.((.------.((((((((((.(........).))))))))))..)).)))......))))))).- ( -26.70, z-score = -2.38, R) >droVir3.scaffold_13047 7524633 96 + 19223366 UUUCAACAAUUUUGGCUGGCCAAGUUGCCGCAAA---CC-AACGCC------AGCCGCAAUAUCAUAAAUAUUUUAUGUUGCGGCAUGUUUUGUAAUUCGUUUGUG-- ......((((((.((....)))))))).((((((---((-((.((.------.((((((((((.(........).))))))))))..)).)))......)))))))-- ( -25.80, z-score = -1.45, R) >consensus UUGCAACCACUGCCUUUGACCAA_UCGCCGAAAAAUGCUCGGCACCGCAGGAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGCUUCUUUU___AUUUGAUUCAAAG ......................................................((((((...(((((.....)))))))))))........................ (-10.10 = -9.50 + -0.60)
| Location | 4,366,282 – 4,366,386 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.87 |
| Shannon entropy | 0.64756 |
| G+C content | 0.41247 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.91 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4366282 104 - 27905053 CUUUGAAUCAAAU---AAAAGAAGCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCCUGCGGUGCCGAGCAUUUUUCGGCGA-UUGGUCAAAGGCAGUGGUUGCAA ((((((.((((..---....(((((((((((((((.....)))))...))))))))))......(((((((.....))))))).-))))))))))((((...)))).. ( -35.60, z-score = -2.88, R) >droSim1.chr3R 10609528 104 + 27517382 CUUUGAAUCUAAU---AAAAGAAGCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCCUGCGGUGCCGAGCAUUUUUCGGCGA-UUGGUCAAAGGCAGUGGUUGCAA ((((((..(((((---....(((((((((((((((.....)))))...))))))))))......(((((((.....))))))))-))))))))))((((...)))).. ( -35.30, z-score = -2.80, R) >droSec1.super_0 3646920 104 + 21120651 CUUUGAAUCUAAU---AAAAGAAGCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGCUUCCUGCGGUGCCGAGCAUUUUUCGGCGA-UUGGUCAAAGGCAGUGGUUGCAA ((((((..(((((---....(((((((((((((((.....)))))...))))))))))......(((((((.....))))))))-))))))))))((((...)))).. ( -37.70, z-score = -3.18, R) >droYak2.chr3R 8439341 104 - 28832112 CUUUGAAUCAAAU---AAAAGAAGCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCCUGCGGUGCCGAGCAUUGUUCGGCGA-UUGGUCAAAGGCAGUGGUUGCCA .(((((.((((..---....(((((((((((((((.....)))))...))))))))))......((((((((...)))))))).-)))))))))(((((...))))). ( -38.30, z-score = -3.42, R) >droEre2.scaffold_4770 536724 104 + 17746568 CUUUGAAUCAAAU---AAAAGAAGCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCCUGCGGUGCCGAGCAUUUUUCGGCGA-UUGGUCAAAGGCAGUGGUUGCAA ((((((.((((..---....(((((((((((((((.....)))))...))))))))))......(((((((.....))))))).-))))))))))((((...)))).. ( -35.60, z-score = -2.88, R) >droAna3.scaffold_13340 19661249 104 + 23697760 CGUGGAAUCAAAU---AAAGAAAGCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUUGAGCGGUGCAGCACAUUUUUCGGCGA-UUGGUCAAAGGCAGUGGCAGUGG (((.(((......---....(((((((((.(((((.....)))))....)))))))))..((......))......))).))).-...((((.......))))..... ( -23.10, z-score = 0.40, R) >dp4.chr2 23335962 93 - 30794189 --CCAUAUCAAAU--AAAAGGAAGCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUGGUGCGGUGCAAAAUAUUUUUCGGCGA-UUGGUCAAAGGCA---------- --...........--........(((((.....(((((((((.((...(((((......)))))..)))))))))))...))((-....))...))).---------- ( -18.40, z-score = 0.25, R) >droPer1.super_3 6108366 93 - 7375914 --CCAUAUCAAAU--AAAAGGAAGCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUGGUGCGGUGCAAAAUAUUUUUCGGCGA-UUGGUCAAAGGCA---------- --...........--........(((((.....(((((((((.((...(((((......)))))..)))))))))))...))((-....))...))).---------- ( -18.40, z-score = 0.25, R) >droWil1.scaffold_181089 926722 100 - 12369635 ------ACUCAGC-GAAAAACAAGCCACAAUCUAAAAUAUUUAUAUUUUUGCGGUAGCCAAACAAAGUUUACAAAAUACAUAAA-ACGGUCAAAAGGAUAGAUGGAUG ------.....((-.........))............(((((((.(((((((.((.............................-)).).)))))).))))))).... ( -6.95, z-score = 1.75, R) >droMoj3.scaffold_6540 3478432 97 - 34148556 -ACACAAACGAAUUACAAAACAUGCCGCAACAUAAAAUAUUUAUGAUAUUGCGGCU------GGCGUUGGG---UUUGCGGCAACUUGGCUAGCCAAAA-AGUUGAAA -.(.(((((......(((..((.((((((((((((.....)))))...))))))))------)...))).)---)))).).((((((((....)))...-)))))... ( -25.80, z-score = -1.55, R) >droVir3.scaffold_13047 7524633 96 - 19223366 --CACAAACGAAUUACAAAACAUGCCGCAACAUAAAAUAUUUAUGAUAUUGCGGCU------GGCGUU-GG---UUUGCGGCAACUUGGCCAGCCAAAAUUGUUGAAA --....(((((............((((((((((((.....)))))...)))))))(------(((..(-((---((...(....)..)))))))))...))))).... ( -26.70, z-score = -1.51, R) >consensus CUUUGAAUCAAAU___AAAAGAAGCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUCCUGCGGUGCCGAGCAUUUUUCGGCGA_UUGGUCAAAGGCAGUGGUUGCAA .......................((((((((((((.....)))))...)))))))........................(((......)))................. (-13.00 = -12.91 + -0.09)
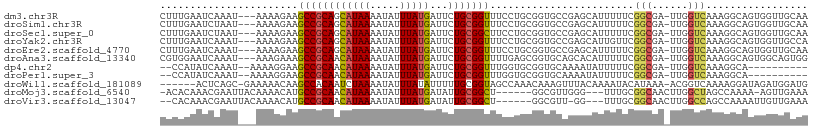

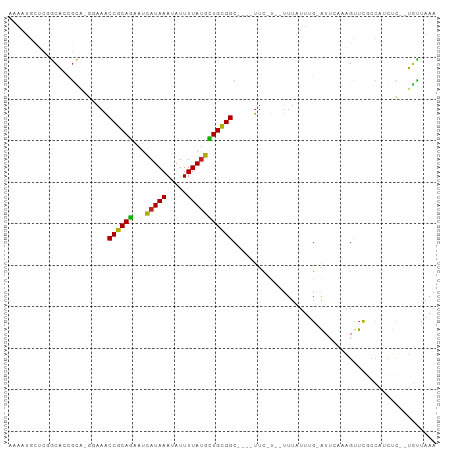
| Location | 4,366,312 – 4,366,404 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.54752 |
| G+C content | 0.37101 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.54 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4366312 92 + 27905053 AAAAUGCUCGGCACCGCA-GGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGC----UUC-U--UUUAUUUG-AUUCAAAGUUCGCCAUCUC--UGUUAAA .........(((.....(-((((.((((((...(((((.....))))))))))).----)))-)--)......(-(........))))).....--....... ( -20.70, z-score = -1.29, R) >droSim1.chr3R 10609558 92 - 27517382 AAAAUGCUCGGCACCGCA-GGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGC----UUC-U--UUUAUUAG-AUUCAAAGUUCGCCAGCUC--UGUUAAA .....(((.(((.....(-((((.((((((...(((((.....))))))))))).----)))-)--)......(-(........))))))))..--....... ( -23.90, z-score = -1.85, R) >droSec1.super_0 3646950 92 - 21120651 AAAAUGCUCGGCACCGCA-GGAAGCCGCAGAAUCAUAAAUAUUUUAUGCUGCGGC----UUC-U--UUUAUUAG-AUUCAAAGUUUGCCAGCUC--UGUUAAA .....(((.(((.....(-(((((((((((...(((((.....))))))))))))----)))-)--).....((-((.....))))))))))..--....... ( -29.10, z-score = -2.95, R) >droYak2.chr3R 8439371 92 + 28832112 ACAAUGCUCGGCACCGCA-GGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGC----UUC-U--UUUAUUUG-AUUCAAAGUUCGCCAUCUC--UGUUAAA .........(((.....(-((((.((((((...(((((.....))))))))))).----)))-)--)......(-(........))))).....--....... ( -20.70, z-score = -1.29, R) >droEre2.scaffold_4770 536754 92 - 17746568 AAAAUGCUCGGCACCGCA-GGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGC----UUC-U--UUUAUUUG-AUUCAAAGUUCGCCAUCUC--UGUUAAA .........(((.....(-((((.((((((...(((((.....))))))))))).----)))-)--)......(-(........))))).....--....... ( -20.70, z-score = -1.29, R) >droAna3.scaffold_13340 19661279 92 - 23697760 AAAAUGUGCUGCACCGCU-CAAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGC----UUU-C--UUUAUUUG-AUUCCACGCUCGGUCCCUC--UAUUAAA ............((((..-(....((((((...(((((.....))))))))))).----..(-(--.......)-)......)..)))).....--....... ( -12.00, z-score = 0.60, R) >dp4.chr2 23335982 89 + 30794189 AAAAUAUUUUGCACCGCA-CCAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGC----UUCCU--UUUAUUUG-AU--AUGGUUCUCUG--UU--UGCUAAA ..........(((..(.(-(((..((((((...(((((.....))))))))))).----...(.--.......)-..--.)))).)..))--).--....... ( -12.00, z-score = 0.38, R) >droPer1.super_3 6108386 89 + 7375914 AAAAUAUUUUGCACCGCA-CCAAACCGCAGAAUCAUAAAUAUUUUAUGUUGCGGC----UUCCU--UUUAUUUG-AU--AUGGUUCUCUG--UU--UGCUAAA ..........(((..(.(-(((..((((((...(((((.....))))))))))).----...(.--.......)-..--.)))).)..))--).--....... ( -12.00, z-score = 0.38, R) >droWil1.scaffold_181089 926761 85 + 12369635 ----------ACUUUGUUUGGCUACCGCAAAAAUAUAAAUAUUUUAGAUUGUGGC----UUGUU--UUUCGCUGAGUU--UCGCUCUUUUGCUUUUGGUUAAA ----------......(((((((((((((((((((....)))))....)))))).----.....--....((.(((..--...)))....))...)))))))) ( -13.70, z-score = -0.04, R) >droVir3.scaffold_13047 7524664 92 + 19223366 ----------AAACCAAC-GCCAGCCGCAAUAUCAUAAAUAUUUUAUGUUGCGGCAUGUUUUGUAAUUCGUUUGUGUUUGCAGCUCAAUUUUUUCUUUUUAAA ----------........-....((((((((((.(........).)))))))))).((..((((((...........))))))..))................ ( -18.20, z-score = -1.41, R) >consensus AAAAUGCUCGGCACCGCA_GGAAACCGCAGAAUCAUAAAUAUUUUAUGCUGCGGC____UUC_U__UUUAUUUG_AUUCAAAGUUCGCCAUCUC__UGUUAAA ........................((((((...(((((.....)))))))))))................................................. (-10.12 = -9.54 + -0.58)
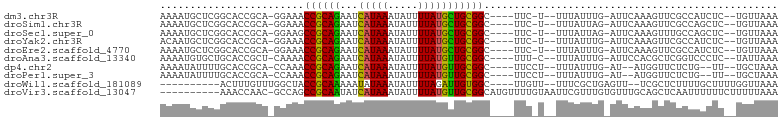
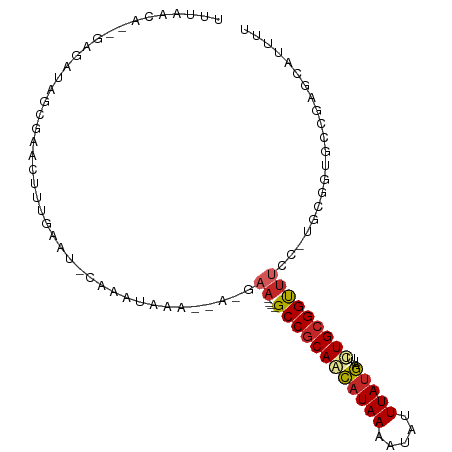
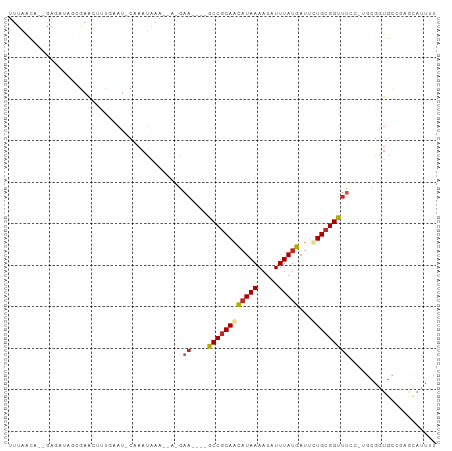
| Location | 4,366,312 – 4,366,404 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.54752 |
| G+C content | 0.37101 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -10.83 |
| Energy contribution | -10.79 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4366312 92 - 27905053 UUUAACA--GAGAUGGCGAACUUUGAAU-CAAAUAAA--A-GAA----GCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCC-UGCGGUGCCGAGCAUUUU .....((--(((........)))))...-........--.-(((----((((((((((((.....)))))...)))))))))).-(((........))).... ( -23.10, z-score = -1.40, R) >droSim1.chr3R 10609558 92 + 27517382 UUUAACA--GAGCUGGCGAACUUUGAAU-CUAAUAAA--A-GAA----GCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCC-UGCGGUGCCGAGCAUUUU .......--..((((((((........)-).......--.-(((----((((((((((((.....)))))...)))))))))).-......))).)))..... ( -25.70, z-score = -1.86, R) >droSec1.super_0 3646950 92 + 21120651 UUUAACA--GAGCUGGCAAACUUUGAAU-CUAAUAAA--A-GAA----GCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGCUUCC-UGCGGUGCCGAGCAUUUU .......--..(((((((....(((...-.)))....--.-(((----((((((((((((.....)))))...)))))))))).-.....)))).)))..... ( -27.50, z-score = -2.40, R) >droYak2.chr3R 8439371 92 - 28832112 UUUAACA--GAGAUGGCGAACUUUGAAU-CAAAUAAA--A-GAA----GCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCC-UGCGGUGCCGAGCAUUGU .....((--(((........)))))...-........--.-(((----((((((((((((.....)))))...)))))))))).-.(((((((...))))))) ( -26.10, z-score = -2.26, R) >droEre2.scaffold_4770 536754 92 + 17746568 UUUAACA--GAGAUGGCGAACUUUGAAU-CAAAUAAA--A-GAA----GCCGCAGCAUAAAAUAUUUAUGAUUCUGCGGUUUCC-UGCGGUGCCGAGCAUUUU .....((--(((........)))))...-........--.-(((----((((((((((((.....)))))...)))))))))).-(((........))).... ( -23.10, z-score = -1.40, R) >droAna3.scaffold_13340 19661279 92 + 23697760 UUUAAUA--GAGGGACCGAGCGUGGAAU-CAAAUAAA--G-AAA----GCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUUG-AGCGGUGCAGCACAUUUU .......--(..(.((((..(.((....-))......--.-(((----((((((.(((((.....)))))....))))))))))-..)))).)..)....... ( -20.90, z-score = -1.19, R) >dp4.chr2 23335982 89 - 30794189 UUUAGCA--AA--CAGAGAACCAU--AU-CAAAUAAA--AGGAA----GCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUGG-UGCGGUGCAAAAUAUUUU ....(((--..--..(...(((..--.(-(.......--.))((----((((((.(((((.....)))))....))))))))))-).)..))).......... ( -17.20, z-score = -1.06, R) >droPer1.super_3 6108386 89 - 7375914 UUUAGCA--AA--CAGAGAACCAU--AU-CAAAUAAA--AGGAA----GCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUGG-UGCGGUGCAAAAUAUUUU ....(((--..--..(...(((..--.(-(.......--.))((----((((((.(((((.....)))))....))))))))))-).)..))).......... ( -17.20, z-score = -1.06, R) >droWil1.scaffold_181089 926761 85 - 12369635 UUUAACCAAAAGCAAAAGAGCGA--AACUCAGCGAAA--AACAA----GCCACAAUCUAAAAUAUUUAUAUUUUUGCGGUAGCCAAACAAAGU---------- ....(((..........(((...--..))).((((((--(....----......................)))))))))).............---------- ( -8.77, z-score = -0.05, R) >droVir3.scaffold_13047 7524664 92 - 19223366 UUUAAAAAGAAAAAAUUGAGCUGCAAACACAAACGAAUUACAAAACAUGCCGCAACAUAAAAUAUUUAUGAUAUUGCGGCUGGC-GUUGGUUU---------- ........................((((...((((..........((.((((((((((((.....)))))...))))))))).)-))).))))---------- ( -17.00, z-score = -0.93, R) >consensus UUUAACA__GAGAUAGCGAACUUUGAAU_CAAAUAAA__A_GAA____GCCGCAACAUAAAAUAUUUAUGAUUCUGCGGUUUCC_UGCGGUGCCGAGCAUUUU ................................................((((((((((((.....)))))...)))))))....................... (-10.83 = -10.79 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:38 2011