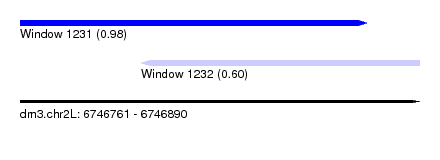
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,746,761 – 6,746,890 |
| Length | 129 |
| Max. P | 0.979623 |

| Location | 6,746,761 – 6,746,873 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Shannon entropy | 0.25274 |
| G+C content | 0.34474 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6746761 112 + 23011544 AAUUAAUUCUGGGCGAGAGGUUGCAAAAUAUUUUUGAUUGUGUUCAAUUAAGAGGCGAAGGUGCAAUGAACCUUUCGCGAUAAUUGAAAAAAGGGAAACAAAAUAAUGCAGU-- ........(((.((((((((((((......((((((((((....))))))))))((....)))))....)))))))))...............(....).........))).-- ( -28.60, z-score = -2.90, R) >droEre2.scaffold_4929 15664242 105 + 26641161 AAUUAAUUCUCG-CGAGAGGUUGCAAAAUCUCGUUGAUUGUGUUCAAUUA-GAGGCGAAGGUGCAAUGAACCUUUCGAGAUAAUUGGAAAAAGGGAAAAUACGGUAU------- ..((((((.((.-(((((((((((....(((.(((((......))))).)-)).((....)))))....)))))))).)).))))))....................------- ( -25.60, z-score = -1.63, R) >droYak2.chr2L 16161205 110 - 22324452 AAUUAAUUAUCG-CGAGGGGUUGCAAAAUAUUUUUGAUUGUGUUCAAUUAGGAGGCGAAGGUGCAAUGAACCUUUCGAGAUAAUUGUAAAAGAGGGAAAACCAUACAGAAU--- ...((((((((.-(((((((((((......((((((((((....))))))))))((....)))))....)))))))).)))))))).......((.....)).........--- ( -30.10, z-score = -3.89, R) >droSec1.super_3 2285671 112 + 7220098 AAUUAAUUCUGG-CGAGAGGUUGCAAAAUAUUUUUGAUUGUGUUCAAUUAAGAGGCGAAGGUGCAAUGAACCUUUCGAGAUAAUUGAAAAAAGGGAAA-AAAAUAAUGCAGUAU ..((((((.(..-(((((((((((......((((((((((....))))))))))((....)))))....))))))))..).))))))...........-............... ( -24.20, z-score = -2.11, R) >droSim1.chr2L 6546827 113 + 22036055 AAUUAAUUCUGG-CGAGAGGUUGCAAAAUAUUUUUGAUUGUGUUCAAUUAAGAGGCGAAGGUGCAAUGAACCUCUUGAGAUAAUUGAAAAAAAAGGGAAAAAAUAAUGCAGUAU ..((((((.(..-(((((((((((......((((((((((....))))))))))((....)))))....))))))))..).))))))........................... ( -24.70, z-score = -2.26, R) >consensus AAUUAAUUCUGG_CGAGAGGUUGCAAAAUAUUUUUGAUUGUGUUCAAUUAAGAGGCGAAGGUGCAAUGAACCUUUCGAGAUAAUUGAAAAAAGGGAAAAAAAAUAAUGCAGU__ ..((((((.....(((((((((((......((((((((((....))))))))))((....)))))....))))))))....))))))........................... (-20.82 = -20.90 + 0.08)

| Location | 6,746,800 – 6,746,890 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Shannon entropy | 0.38068 |
| G+C content | 0.38201 |
| Mean single sequence MFE | -12.16 |
| Consensus MFE | -7.42 |
| Energy contribution | -7.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6746800 90 - 23011544 -UUUGGCCCUCUUAG--UUGACUGCAUUAUUUUGUUUCCCUUUUUUCAAUUAUCGCGAAAGGUUCAUUGCACCUUCGCCUCUUAAUUGAACAC -....((..((....--..))..))...................((((((((..(((((.(((.......))))))))....))))))))... ( -16.20, z-score = -1.86, R) >droEre2.scaffold_4929 15664280 85 - 26641161 CGUUUACCCACAGAACGAAUACCGUA-----UUUUCCC--UUUUUCCAAUUAUCUCGAAAGGUUCAUUGCACCUUCGCCUC-UAAUUGAACAC (((((.......))))).........-----.......--......((((((...((((.(((.......)))))))....-))))))..... ( -10.50, z-score = -0.98, R) >droYak2.chr2L 16161243 92 + 22324452 CAUUCGCCCUCUAAG-AAUGAAUUCUGUAUGGUUUUCCCUCUUUUACAAUUAUCUCGAAAGGUUCAUUGCACCUUCGCCUCCUAAUUGAACAC ............(((-(..(((..((....))..)))..))))...((((((...((((.(((.......))))))).....))))))..... ( -11.40, z-score = -0.06, R) >droSec1.super_3 2285709 90 - 7220098 --UUUACCCACACAACGAAUACUGCAUUAUUUUUUUCC-CUUUUUUCAAUUAUCUCGAAAGGUUCAUUGCACCUUCGCCUCUUAAUUGAACAC --....................................-.....((((((((...((((.(((.......))))))).....))))))))... ( -10.20, z-score = -2.26, R) >droSim1.chr2L 6546865 91 - 22036055 --UUUACCCACAGAACGAAUACUGCAUUAUUUUUUCCCUUUUUUUUCAAUUAUCUCAAGAGGUUCAUUGCACCUUCGCCUCUUAAUUGAACAC --..........(((.(((((......))))).)))........((((((((.....((((((.............))))))))))))))... ( -12.52, z-score = -2.56, R) >consensus __UUUACCCACAGAACGAAUACUGCAUUAUUUUUUUCC_CUUUUUUCAAUUAUCUCGAAAGGUUCAUUGCACCUUCGCCUCUUAAUUGAACAC ............................................((((((((...((.(((((.......))))))).....))))))))... ( -7.42 = -7.86 + 0.44)
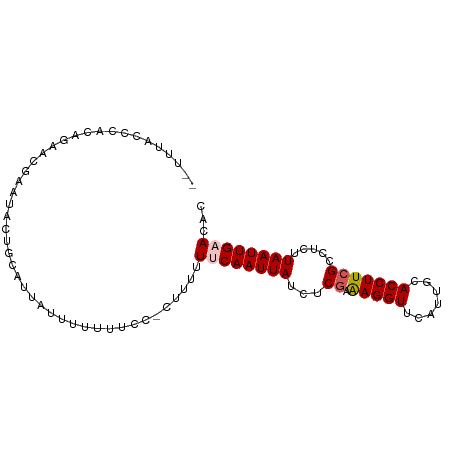
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:51 2011