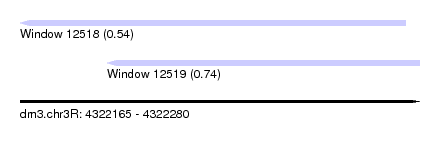
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,322,165 – 4,322,280 |
| Length | 115 |
| Max. P | 0.741513 |

| Location | 4,322,165 – 4,322,276 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Shannon entropy | 0.37242 |
| G+C content | 0.49408 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -10.51 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4322165 111 - 27905053 --------CUUCACUUUCCGACCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUCGACAGUUUUCUUCCCUUUUUGCGCA --------.........................(((((.....((((....)))).((((((.....))))))...........(((.((....)))))..............))))). ( -17.90, z-score = -2.14, R) >droSim1.chr3R 10564322 111 + 27517382 --------CUUCACUUUCCGUCCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUCGACAGUUUUCUUCCCUUUUUGCGCA --------.........................(((((.....((((....)))).((((((.....))))))...........(((.((....)))))..............))))). ( -17.90, z-score = -2.44, R) >droSec1.super_0 3603096 111 + 21120651 --------CUUCACUUUCCGUCCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUCGACAGUUUUCUUCCCUUUUUGCGCA --------.........................(((((.....((((....)))).((((((.....))))))...........(((.((....)))))..............))))). ( -17.90, z-score = -2.44, R) >droYak2.chr3R 8394409 111 - 28832112 --------CUCCACUUUACGUCCCGUUGCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUCGACAGUUUUCUUCCCUUUUUGCGCA --------.........................(((((.....((((....)))).((((((.....))))))...........(((.((....)))))..............))))). ( -17.90, z-score = -1.59, R) >droEre2.scaffold_4770 492603 111 + 17746568 --------CUCCACUUUUCGCCCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUCGACAGUUUUCUUCCCUUUUUGCGCA --------.........................(((((.....((((....)))).((((((.....))))))...........(((.((....)))))..............))))). ( -17.90, z-score = -2.22, R) >droAna3.scaffold_13340 19617762 103 + 23697760 ----------------UUCCCCGACCAUCCUUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCUUCCCUUUUUGCGCA ----------------.................(((((.....((((....)))).((((((.....))))))...........((((......).)))..............))))). ( -16.40, z-score = -1.90, R) >dp4.chr2 23289641 96 - 30794189 -----------------------UUGCCCCUUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCUUCGCUUUUUGCGCA -----------------------.(((....(((.((......((((....)))).((((((.....)))))).................)).)))...........((.....))))) ( -16.50, z-score = -0.60, R) >droPer1.super_3 6061951 96 - 7375914 -----------------------UUGCCCCUUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCUUCGCUUUUUGCGCA -----------------------.(((....(((.((......((((....)))).((((((.....)))))).................)).)))...........((.....))))) ( -16.50, z-score = -0.60, R) >droVir3.scaffold_13047 7474656 116 - 19223366 CCUCCCCUCCAGCCCCGCCACGCCUCAUCCCGCUCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCC---UUUGCUGCGCA ...........((...((...((........))..(((.....((((....)))).((((((.....))))))............)))..))..(.((((.....---...))))))). ( -17.20, z-score = -0.75, R) >droMoj3.scaffold_6540 3408559 94 - 34148556 ----------------------CCCCAUCCCGUUCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCC---UUUGCUGCGCG ----------------------.............(((.....((((....)))).((((((.....))))))............))).(((..(.((((.....---...)))))))) ( -15.30, z-score = -1.27, R) >droGri2.scaffold_14830 3330295 94 - 6267026 -----------------------CCCAUCCUGCUCGCAUUUCCGUUGCCAUUAGCCAACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCC--UUUUGCCGCGCA -----------------------.......(((..(((.......))).........(((((.....)))))..............))).((.((.(((......--..))).)).)). ( -10.10, z-score = 0.45, R) >consensus ________C____C__U_CG_CCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUCGACAGUUUUCUUCCCUUUUUGCGCA ...................................(((.....((((....)))).((((((.....))))))............))).(((......................))).. (-10.51 = -10.52 + 0.01)



| Location | 4,322,190 – 4,322,280 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Shannon entropy | 0.41569 |
| G+C content | 0.52285 |
| Mean single sequence MFE | -12.60 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4322190 90 - 27905053 ---------------------------CUGCCUUCA--CUUUCCGACCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUC ---------------------------.........--...................((.(((.....((((....)))).((((((.....))))))............))).))... ( -11.20, z-score = -0.90, R) >droSim1.chr3R 10564347 90 + 27517382 ---------------------------CGGCCUUCA--CUUUCCGUCCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUC ---------------------------(((......--....)))............((.(((.....((((....)))).((((((.....))))))............))).))... ( -11.50, z-score = -0.46, R) >droYak2.chr3R 8394434 90 - 28832112 ---------------------------CUGCCUCCA--CUUUACGUCCCGUUGCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUC ---------------------------..((.....--..............((....))(((.....((((....)))).((((((.....))))))............)))..)).. ( -11.30, z-score = -0.39, R) >droEre2.scaffold_4770 492628 90 + 17746568 ---------------------------CUGCCUCCA--CUUUUCGCCCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCUCGCUC ---------------------------.........--...................((.(((.....((((....)))).((((((.....))))))............))).))... ( -11.20, z-score = -1.01, R) >droAna3.scaffold_13340 19617787 81 + 23697760 --------------------------------------CUGUUCCCCGACCAUCCUUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUC --------------------------------------...................((.(((.....((((....)))).((((((.....))))))............))).))... ( -11.60, z-score = -1.50, R) >dp4.chr2 23289666 75 - 30794189 --------------------------------------------GUGGUUGCCCCUUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUC --------------------------------------------(((...((......))(((.....((((....)))).((((((.....))))))............))).))).. ( -13.20, z-score = -0.80, R) >droPer1.super_3 6061976 75 - 7375914 --------------------------------------------GUGGUUGCCCCUUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUC --------------------------------------------(((...((......))(((.....((((....)))).((((((.....))))))............))).))).. ( -13.20, z-score = -0.80, R) >droVir3.scaffold_13047 7474678 119 - 19223366 CUCUCGGUCUCGGUCUCGCUCUCGCCCUCCCCUCCAGCCCCGCCACGCCUCAUCCCGCUCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUC .....(((...(((...(((...............)))...)))..))).......((..(((.....((((....)))).((((((.....))))))............)))..)).. ( -17.56, z-score = -0.36, R) >consensus ___________________________C_GCCU_CA__CUUUCCGUCCCGUUCCCAUCGCGCAUUUCCGCUGCCAUUAGCCGACAAACUUCAUUUGUCAUCUCAUUUUUCUGCACGCUC .........................................................((.(((.....((((....)))).((((((.....))))))............))).))... (-10.80 = -10.92 + 0.12)

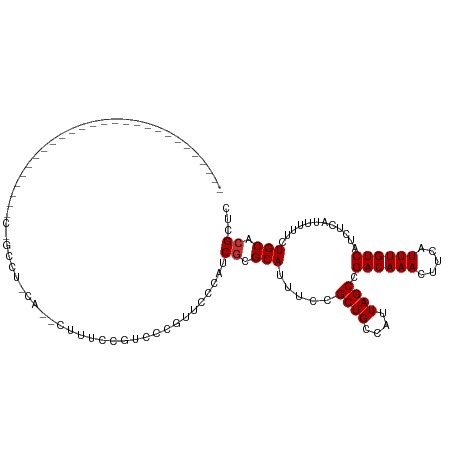

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:29 2011