
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,304,765 – 4,304,869 |
| Length | 104 |
| Max. P | 0.529680 |

| Location | 4,304,765 – 4,304,869 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.61 |
| Shannon entropy | 0.56747 |
| G+C content | 0.56568 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -10.39 |
| Energy contribution | -10.64 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
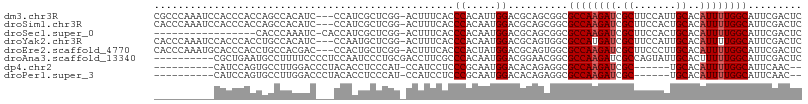
>dm3.chr3R 4304765 104 - 27905053 CGCCCAAAUCCACCCACCAGCCACAUC---CCAUCGCUCGG-ACUUUCACCCACAUUGGACGCAGCGGCGCCAAGAUCGCUUCCAUUGCACAUUUUGGCAUUCGACUC .((((...........(((((......---.....))).))-........((.....)).....).)))((((((((.((.......))..))))))))......... ( -20.60, z-score = -0.25, R) >droSim1.chr3R 10546779 104 + 27517382 CACCCAAAUCCACCCACCAGCCACAUC---CCAUCGCUCGG-ACUUUCACCCACAAUGGACGCAGCGGCGCCAAGAUCGCUUCCACUGCACAUUUUGGCAUUCGACUC ...........................---...(((((((.-.(.............)..)).))))).((((((((.((.......))..))))))))......... ( -19.72, z-score = -0.60, R) >droSec1.super_0 3585759 89 + 21120651 -----------------CACCCAAAUC-CACCAUCGCUCGG-ACUUUCACCCACAAUGGACGCAGCGGCGCCAAGAUCGCUUCCACUGCACAUUUUGGCAUUCGACUC -----------------..........-.....(((((((.-.(.............)..)).))))).((((((((.((.......))..))))))))......... ( -19.72, z-score = -1.21, R) >droYak2.chr3R 8376414 104 - 28832112 CACCCAAAUCCACCCACCUGCCACAUC---CCAAUGCUCGG-ACUUUCACCCACAAUGGACGCAGUGGCGCCAUGAUCGCUUCCAUUGCACAUUUUGGCAUUCGACUC ..................(((((....---(((.((...((-........)).)).)))..(((((((.((.......))..)))))))......)))))........ ( -21.50, z-score = -0.59, R) >droEre2.scaffold_4770 474083 104 + 17746568 CACCCAAAUGCACCCACCUGCCACGAC---CCACUGCUCGG-ACUUUCACCCACUAUGGACGCAGUGGCGCCAAGAUCGCUUCCCUUGCACAUUUUGGCAUUCGACUC .........(((......)))..(((.---(((((((...(-.....)..((.....))..))))))).((((((((.((.......))..))))))))..))).... ( -26.20, z-score = -2.20, R) >droAna3.scaffold_13340 4092627 98 - 23697760 ----------CGCUGAAUGCCUUUUCCCCUCCAAUCCCUGCGACCUUCGCCCACAAUGGACGGAACGGCGCCAAGAUCGCCAGUAUUGCACUUUUUGGCAUUCGACUC ----------...((((((((.....((.((((......(((.....)))......)))).))...((((.......))))...............)))))))).... ( -27.60, z-score = -2.05, R) >dp4.chr2 7519128 89 + 30794189 ----------CAUCCAGUGCCUUGGACCCUACACCUCCCAU-CCAUCCUCCCGCAAUGGACACAGAGGCGCCAAGAUCGC------UGCACAUUUUGGCAUUCAAC-- ----------..(((((....))))).......((((...(-((((.........)))))....)))).((((((((...------.....)))))))).......-- ( -20.60, z-score = -0.95, R) >droPer1.super_3 1328396 89 + 7375914 ----------CAUCCAGUGCCUUGGACCCUACACCUCCCAU-CCAUCCUCCCGCAAUGGACACAGAGGCGCCAAGAUCGC------UGCACAUUUUGGCAUUCAAC-- ----------..(((((....))))).......((((...(-((((.........)))))....)))).((((((((...------.....)))))))).......-- ( -20.60, z-score = -0.95, R) >consensus __________CACCCACCACCCACAUC___CCACCGCUCGG_ACUUUCACCCACAAUGGACGCAGCGGCGCCAAGAUCGCUUCCAUUGCACAUUUUGGCAUUCGACUC ..................................................((.....))..........((((((((.((.......))..))))))))......... (-10.39 = -10.64 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:27 2011