
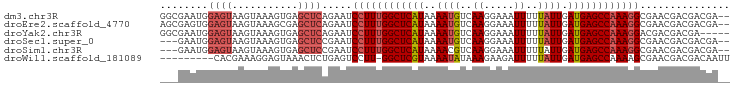
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,302,696 – 4,302,789 |
| Length | 93 |
| Max. P | 0.996211 |

| Location | 4,302,696 – 4,302,789 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.38 |
| Shannon entropy | 0.29708 |
| G+C content | 0.41173 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4302696 93 + 27905053 GGCGAAUGGAGUAAGUAAAGUGAGCUCAGAAUCCUUUGGCUCAUAAAAUGUCAAGGAAAUUUUUAUUGAUGAGCCAAAGGCGAACGACGACGA-- ..((....((((...........)))).....((((((((((((..((((..((.....))..)))).))))))))))))....)).......-- ( -22.00, z-score = -1.15, R) >droEre2.scaffold_4770 471970 93 - 17746568 AGCGAGUGGAGUAAGUAAAGCGAGCUCAGAAUCCUUUGGCUCAUAAAAUGUCAAGGAAAUUUUUAUUGAUGAGCCAAAGGCGAACGACGACGA-- .(((..((((((..(.....)..)))).....((((((((((((..((((..((.....))..)))).))))))))))))))..)).).....-- ( -24.30, z-score = -1.55, R) >droYak2.chr3R 8374270 90 + 28832112 GGCGAAUGGAGUAAGUAAAGUGAGCUCAGAAUCCUUUGGCUCAUAAAAUGUCAAGGAAAUUUUUAUUGAUGAGCCAAAGGACGACGACGA----- ..((....((((...........))))....(((((((((((((..((((..((.....))..)))).))))))))))))))).......----- ( -24.20, z-score = -2.34, R) >droSec1.super_0 3583689 90 - 21120651 ---GAAUGGAGUAAGUAAAGUGAGCUCCGAAUCCUUUGGCUCAUAAAAUGUCAAGGAAAUUUUUAUUGAUGAGCCAAAGGCGAACGACGACGA-- ---...((((((...........))))))...((((((((((((..((((..((.....))..)))).)))))))))))).............-- ( -24.90, z-score = -2.62, R) >droSim1.chr3R 10544724 90 - 27517382 ---GAAUGGAGUAAGUAAAGUGAGCUCCGAAUCCUUUGGCUCAUAAAACGUCAAGGAAAUUUUUAUUGAUGAGCCAAAGGCGAACGACGACGA-- ---...((((((...........))))))...(((((((((((.......((((...........))))))))))))))).............-- ( -23.61, z-score = -1.99, R) >droWil1.scaffold_181089 864353 85 + 12369635 ---------CACGAAAGGAGUAAACUCUGAGUCCUU-GGCUCGUAAAAUAUAAAGAAGAUUUUUAUUGAUGAGCCAAAAGCGAACGACGACAAUU ---------..((...((((....))))..((..((-(((((((.....(((((((...)))))))..)))))))))..))...))......... ( -20.20, z-score = -2.74, R) >consensus ___GAAUGGAGUAAGUAAAGUGAGCUCAGAAUCCUUUGGCUCAUAAAAUGUCAAGGAAAUUUUUAUUGAUGAGCCAAAGGCGAACGACGACGA__ ........((((...........)))).....((((((((((((..((((..((.....))..)))).))))))))))))............... (-16.79 = -17.68 + 0.89)
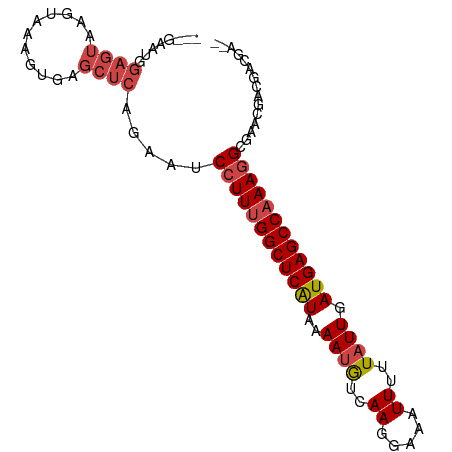
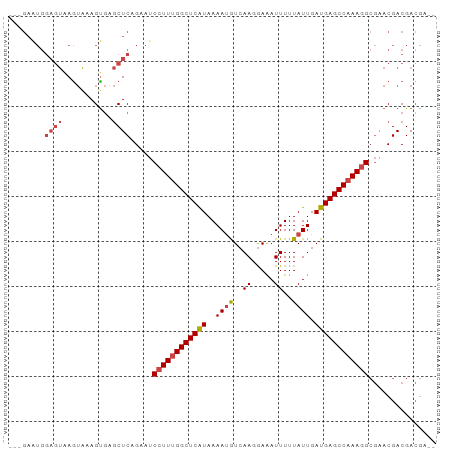
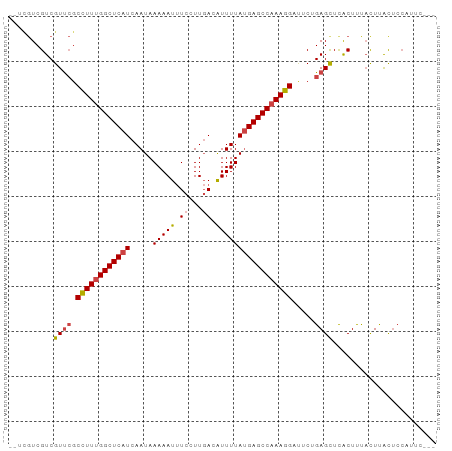
| Location | 4,302,696 – 4,302,789 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Shannon entropy | 0.29708 |
| G+C content | 0.41173 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.996211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4302696 93 - 27905053 --UCGUCGUCGUUCGCCUUUGGCUCAUCAAUAAAAAUUUCCUUGACAUUUUAUGAGCCAAAGGAUUCUGAGCUCACUUUACUUACUCCAUUCGCC --........(((((((((((((((((.....(((((.((...)).)))))))))))))))))....)))))....................... ( -21.80, z-score = -3.12, R) >droEre2.scaffold_4770 471970 93 + 17746568 --UCGUCGUCGUUCGCCUUUGGCUCAUCAAUAAAAAUUUCCUUGACAUUUUAUGAGCCAAAGGAUUCUGAGCUCGCUUUACUUACUCCACUCGCU --..(.((..(((((((((((((((((.....(((((.((...)).)))))))))))))))))....))))).)))................... ( -22.10, z-score = -2.62, R) >droYak2.chr3R 8374270 90 - 28832112 -----UCGUCGUCGUCCUUUGGCUCAUCAAUAAAAAUUUCCUUGACAUUUUAUGAGCCAAAGGAUUCUGAGCUCACUUUACUUACUCCAUUCGCC -----...(((..((((((((((((((.....(((((.((...)).)))))))))))))))))))..)))......................... ( -23.10, z-score = -4.13, R) >droSec1.super_0 3583689 90 + 21120651 --UCGUCGUCGUUCGCCUUUGGCUCAUCAAUAAAAAUUUCCUUGACAUUUUAUGAGCCAAAGGAUUCGGAGCUCACUUUACUUACUCCAUUC--- --.............((((((((((((.....(((((.((...)).)))))))))))))))))....((((.............))))....--- ( -22.92, z-score = -3.50, R) >droSim1.chr3R 10544724 90 + 27517382 --UCGUCGUCGUUCGCCUUUGGCUCAUCAAUAAAAAUUUCCUUGACGUUUUAUGAGCCAAAGGAUUCGGAGCUCACUUUACUUACUCCAUUC--- --.............((((((((((((.....(((((.((...)).)))))))))))))))))....((((.............))))....--- ( -23.22, z-score = -3.18, R) >droWil1.scaffold_181089 864353 85 - 12369635 AAUUGUCGUCGUUCGCUUUUGGCUCAUCAAUAAAAAUCUUCUUUAUAUUUUACGAGCC-AAGGACUCAGAGUUUACUCCUUUCGUG--------- ....(((((.....)).((((((((...((((.(((.....))).))))....)))))-))))))...(((....)))........--------- ( -13.20, z-score = -0.63, R) >consensus __UCGUCGUCGUUCGCCUUUGGCUCAUCAAUAAAAAUUUCCUUGACAUUUUAUGAGCCAAAGGAUUCUGAGCUCACUUUACUUACUCCAUUC___ ..........((((.((((((((((((.....(((((.((...)).))))))))))))))))).....))))....................... (-17.83 = -18.25 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:26 2011