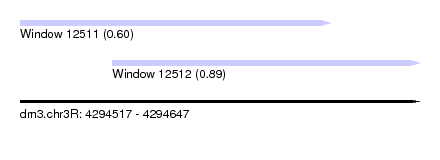
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,294,517 – 4,294,647 |
| Length | 130 |
| Max. P | 0.894099 |

| Location | 4,294,517 – 4,294,618 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Shannon entropy | 0.44215 |
| G+C content | 0.59592 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -21.64 |
| Energy contribution | -23.20 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
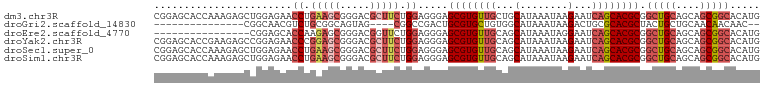
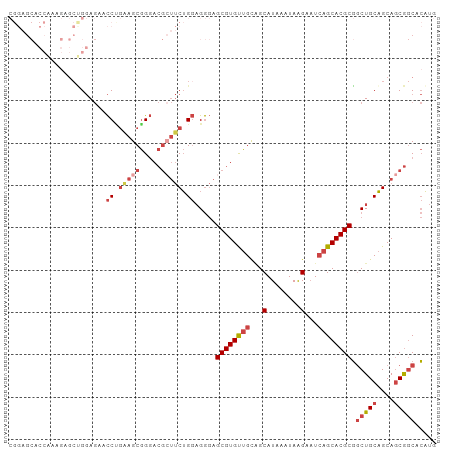
>dm3.chr3R 4294517 101 + 27905053 CGGAGCACCAAAGAGCUGGAGAACCUGAAGCGGGACGCUUCUGGAGGGAGCGUGUUGCUGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUG ....((.(((......)))....((.((((((...)))))).)).....))((((((((((............((((.....))))..))))))))))... ( -33.94, z-score = -0.55, R) >droGri2.scaffold_14830 3281970 80 + 6267026 ---------------CGGCAACGUCUGCGGCAGUAG----CGGCCGACUGCGUGCUGUGGCAUAAAUAAGACUGCGCACGCUACUGCUGCAACAACAAC-- ---------------((....))..(((((((((((----(.((.....))((((.(..((........).)..)))))))))))))))))........-- ( -34.20, z-score = -2.34, R) >droEre2.scaffold_4770 463724 85 - 17746568 ----------------CGGAGCACCAAGAGCGGGACGGUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAGGAAUCAGCACGCGGCUGCAGCAGCGGCACAUG ----------------((((((.((......))....))))))......((((((((...(........)...)))))))).(((((....)))))..... ( -28.60, z-score = -1.27, R) >droYak2.chr3R 8366070 101 + 28832112 CGGAGCACCGAAGAGCCGGAGAACCCGGAGCGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUG ....((.(((.....((((.....))))..)))..((((.(((.((...((((((((...(........)...))))))))..)).))).))))))..... ( -38.30, z-score = -1.72, R) >droSec1.super_0 3575658 101 - 21120651 CGGAGCACCAAAGAGCUGGAGAACCUGAAGCGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUG .((....)).....((((.((..((.((((((...)))))).)).....((((((((...(........)...))))))))..)).))))........... ( -32.30, z-score = -0.31, R) >droSim1.chr3R 10536700 101 - 27517382 CGGAGCACCAAAGAGCUGGAGAACCUGAAGCGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUG .((....)).....((((.((..((.((((((...)))))).)).....((((((((...(........)...))))))))..)).))))........... ( -32.30, z-score = -0.31, R) >consensus CGGAGCACCAAAGAGCUGGAGAACCUGAAGCGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUG .......................((.(((((.....))))).)).....((((((((...(........)...)))))))).(((((....)))))..... (-21.64 = -23.20 + 1.56)
| Location | 4,294,547 – 4,294,647 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.36 |
| Shannon entropy | 0.14841 |
| G+C content | 0.58185 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
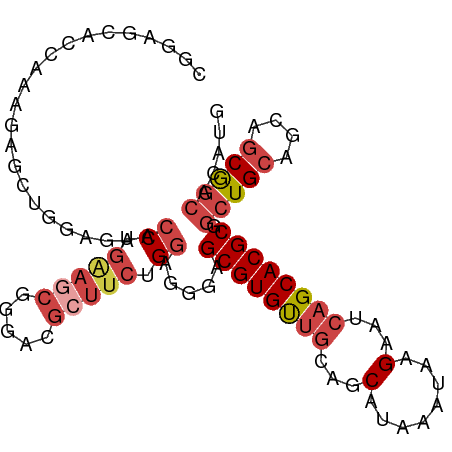
>dm3.chr3R 4294547 100 + 27905053 CGGGACGCUUCUGGAGGGAGCGUGUUGCUGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUGAAAAG----GGAGGACAAGAAGAAGCGGCAGUC .....(((((((....(..((((((((...(........)...)))))))).(((((....)))))..........----......)....)))))))...... ( -31.60, z-score = -1.59, R) >droEre2.scaffold_4770 463738 104 - 17746568 CGGGACGGUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAGGAAUCAGCACGCGGCUGCAGCAGCGGCACAUGAAAAACGGAGAGGGGGGCGAGGAAGCGGCAGCC .....(..(((((......((((((((...(........)...)))))))).(((((....)))))..........)))))..)..(((..(....)....))) ( -28.20, z-score = 0.03, R) >droYak2.chr3R 8366100 101 + 28832112 CGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUGAAAAAC---GAAGGAGAAGGAGGAGGGGGAGUC ...(((.(((((.......((((((((...(........)...)))))))).(((((....)))))...........---...............))))).))) ( -26.90, z-score = -1.41, R) >droSec1.super_0 3575688 100 - 21120651 CGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUGAAAAG----GGAGGACAAGAAGAAGCGGCAGUC .....(((((((....(..((((((((...(........)...)))))))).(((((....)))))..........----......)....)))))))...... ( -31.60, z-score = -2.14, R) >droSim1.chr3R 10536730 100 - 27517382 CGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUGAAAAG----GGAGGACAAGAAGAAGCGGCAGUC .....(((((((....(..((((((((...(........)...)))))))).(((((....)))))..........----......)....)))))))...... ( -31.60, z-score = -2.14, R) >consensus CGGGACGCUUCUGGAGGGAGCGUGUUGCAGCAUAAAUAAGAAUCAGCACGCGGCUGCAGCAGCGGCACAUGAAAAG____GGAGGACAAGAAGAAGCGGCAGUC .....(((((((.......((((((((...(........)...)))))))).(((((....))))).........................)))))))...... (-28.66 = -28.74 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:24 2011