| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,221,862 – 4,221,957 |
| Length | 95 |
| Max. P | 0.633750 |

| Location | 4,221,862 – 4,221,957 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.52471 |
| G+C content | 0.41268 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.63 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
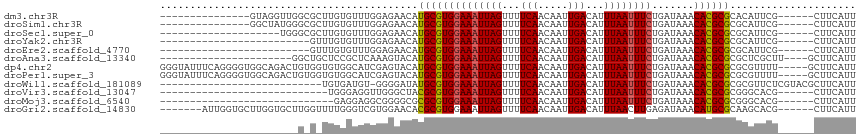
>dm3.chr3R 4221862 95 + 27905053 ---------------GUAGGUUGGCGCUUGUGUUUGGAGAACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCACAUUCG------CUUCAUU ---------------.......((((..(((((.((.....)).(((((((((((((...(((.....)))...)))))))).......))))))))))..))------))..... ( -22.11, z-score = -0.75, R) >droSim1.chr3R 10474209 95 - 27517382 ---------------GGCUAUGGGCGCUUGUGUUUGGAGAACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCAUUCG------CUUCAUU ---------------(((.(((.((((..(((((((.((((..((.(((..((((.((.....))))))..))).))..))))..))))))))))).)))..)------))..... ( -25.00, z-score = -1.44, R) >droSec1.super_0 3512817 90 - 21120651 --------------------UGGGCGCUUGUGUUUGGAGAACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCAUUCG------CUUCAUU --------------------...((((.((((((((.((((..((.(((..((((.((.....))))))..))).))..))))..)))))))).)))).....------....... ( -22.30, z-score = -0.97, R) >droYak2.chr3R 8300543 85 + 28832112 -------------------------GUUUGUGUUUGGAGAACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCAUUCG------CUUCAUU -------------------------.........(((((...(((((((((((((((...(((.....)))...))))))))((.....))..)))))))...------))))).. ( -19.00, z-score = -0.83, R) >droEre2.scaffold_4770 400687 85 - 17746568 -------------------------GUUUGUGUUUGGAGAACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCAUUCG------CUUCAUU -------------------------.........(((((...(((((((((((((((...(((.....)))...))))))))((.....))..)))))))...------))))).. ( -19.00, z-score = -0.83, R) >droAna3.scaffold_13340 19525288 90 - 23697760 ----------------------GGCUGCUCCGCUCAAAGUACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCUCGCUU----GCUUCAUU ----------------------(((......)))..(((((...(((((((((((((...(((.....)))...)))))))).......)))))((....)).)----)))).... ( -16.21, z-score = 0.61, R) >dp4.chr2 23186695 111 + 30794189 GGGUAUUUCAGGGGUGGCAGACUGUGGUGUGGCAUCGAGUACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCGUUUU-----GCUUCAUU .............(.((((((.(((((((((((((.......))))..(((((((((...(((.....)))...)))))))))......))))).)))).)))-----))).)... ( -28.20, z-score = -0.59, R) >droPer1.super_3 5959122 111 + 7375914 GGGUAUUUCAGGGGUGGCAGACUGUGGUGUGGCAUCGAGUACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCGUUUU-----GCUUCAUU .............(.((((((.(((((((((((((.......))))..(((((((((...(((.....)))...)))))))))......))))).)))).)))-----))).)... ( -28.20, z-score = -0.59, R) >droWil1.scaffold_181089 787858 88 + 12369635 ---------------------------UGUGAUGU-GGGGAUAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCGUUCUCGUACGCUUCAUU ---------------------------.(((.(((-(((((..((((((((((((((...(((.....)))...))))))))((.....))))))))..)))))))))))...... ( -22.70, z-score = -1.32, R) >droVir3.scaffold_13047 7347729 82 + 19223366 ----------------------------UGGGAGGUUGGGCUACGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGGGCACG------CUUCAUU ----------------------------...(((((..(.(..((((((((((((((...(((.....)))...)))))))).......))))))..).)..)------))))... ( -23.51, z-score = -2.96, R) >droMoj3.scaffold_6540 3265178 81 + 34148556 -----------------------------GAGGAGGCGGGGCGCGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGGGCACG------CUUCAUU -----------------------------...((((((..((.((((((((((((((...(((.....)))...))))))))((.....)))))))).)).))------))))... ( -30.10, z-score = -4.76, R) >droGri2.scaffold_14830 3189136 103 + 6267026 -------AUUGGUGCUUGGUGCUUGGUUUUGGGUCGUGGAACACGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAACUUGAGAUAAACAUGCGCAAGCACG------CUUCAUU -------....(((((((.(((.((((((..(((((((.....))))((((((....))))))..............)))..))))...)).)))))))))).------....... ( -27.10, z-score = -0.91, R) >consensus _______________________G_G_UUGGGUUUGGAGAACAUGCGUGGAAAUUAGUUUUCAACAAUUGACAUUUAAUUUCUGAUAAACACGCGCGCGUUCG______CUUCAUU ...........................................((((((((((((((...(((.....)))...)))))))).......))))))..................... (-11.72 = -11.63 + -0.10)

| Location | 4,221,862 – 4,221,957 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.52471 |
| G+C content | 0.41268 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -10.89 |
| Energy contribution | -10.98 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4221862 95 - 27905053 AAUGAAG------CGAAUGUGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUUCUCCAAACACAAGCGCCAACCUAC--------------- ......(------((..((((.(((((.......(((((((....(((.....)))....)))))))))))).((.....))..))))..)))........--------------- ( -18.51, z-score = -1.03, R) >droSim1.chr3R 10474209 95 + 27517382 AAUGAAG------CGAAUGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUUCUCCAAACACAAGCGCCCAUAGCC--------------- ......(------(....(((((((((.......(((((((....(((.....)))....)))))))))))).((((.....))))...)))).....)).--------------- ( -20.81, z-score = -1.55, R) >droSec1.super_0 3512817 90 + 21120651 AAUGAAG------CGAAUGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUUCUCCAAACACAAGCGCCCA-------------------- .......------.....(((((((((.......(((((((....(((.....)))....)))))))))))).((((.....))))...))))...-------------------- ( -19.01, z-score = -1.03, R) >droYak2.chr3R 8300543 85 - 28832112 AAUGAAG------CGAAUGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUUCUCCAAACACAAAC------------------------- ...((((------((....)))(((((.......(((((((....(((.....)))....))))))))))))...))).............------------------------- ( -15.41, z-score = -0.53, R) >droEre2.scaffold_4770 400687 85 + 17746568 AAUGAAG------CGAAUGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUUCUCCAAACACAAAC------------------------- ...((((------((....)))(((((.......(((((((....(((.....)))....))))))))))))...))).............------------------------- ( -15.41, z-score = -0.53, R) >droAna3.scaffold_13340 19525288 90 + 23697760 AAUGAAGC----AAGCGAGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUACUUUGAGCGGAGCAGCC---------------------- ......((----..((....))(((((.......(((((((....(((.....)))....))))))))))))...........)).........---------------------- ( -15.91, z-score = 1.52, R) >dp4.chr2 23186695 111 - 30794189 AAUGAAGC-----AAAACGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUACUCGAUGCCACACCACAGUCUGCCACCCCUGAAAUACCC ......((-----(....(((.(((((.......(((((((....(((.....)))....)))))))))))).)))....(((...........))))))................ ( -14.71, z-score = 0.82, R) >droPer1.super_3 5959122 111 - 7375914 AAUGAAGC-----AAAACGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUACUCGAUGCCACACCACAGUCUGCCACCCCUGAAAUACCC ......((-----(....(((.(((((.......(((((((....(((.....)))....)))))))))))).)))....(((...........))))))................ ( -14.71, z-score = 0.82, R) >droWil1.scaffold_181089 787858 88 - 12369635 AAUGAAGCGUACGAGAACGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUAUCCCC-ACAUCACA--------------------------- ..(((.((((......))))..(((((.......(((((((....(((.....)))....))))))))))))........-...)))..--------------------------- ( -17.41, z-score = -1.17, R) >droVir3.scaffold_13047 7347729 82 - 19223366 AAUGAAG------CGUGCCCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCGUAGCCCAACCUCCCA---------------------------- .......------.(.((..(((((((.......(((((((....(((.....)))....)))))))))))))).)).).........---------------------------- ( -16.91, z-score = -2.08, R) >droMoj3.scaffold_6540 3265178 81 - 34148556 AAUGAAG------CGUGCCCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCGCGCCCCGCCUCCUC----------------------------- ...((.(------((.(..((((((((.......(((((((....(((.....)))....)))))))))))))))..)))).))...----------------------------- ( -24.21, z-score = -4.24, R) >droGri2.scaffold_14830 3189136 103 - 6267026 AAUGAAG------CGUGCUUGCGCAUGUUUAUCUCAAGUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCGUGUUCCACGACCCAAAACCAAGCACCAAGCACCAAU------- ......(------.(((((((.((..(((((........)))))(((....((.((((.....)))).))..(((...))))))..........))..))))))))...------- ( -18.20, z-score = -0.40, R) >consensus AAUGAAG______CGAACGCGCGCGUGUUUAUCAGAAAUUAAAUGUCAAUUGUUGAAAACUAAUUUCCACGCAUGUUCUCCAAACACAA_C_C_______________________ ......................(((((.......(((((((....(((.....)))....))))))))))))............................................ (-10.89 = -10.98 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:17 2011