
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,213,667 – 4,213,766 |
| Length | 99 |
| Max. P | 0.819084 |

| Location | 4,213,667 – 4,213,766 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 64.14 |
| Shannon entropy | 0.80512 |
| G+C content | 0.58541 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -11.39 |
| Energy contribution | -11.25 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
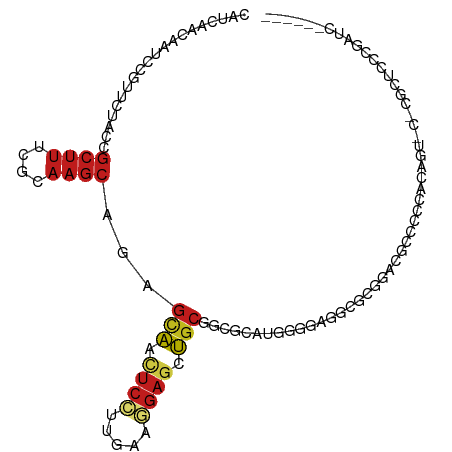
>dm3.chr3R 4213667 99 + 27905053 ------GAUCGCGAGCG-G-ACUGAUGGAGCAUCCGAGCCCGUCU-UGCGCCGCAGCUCCUUCAAGGAGUUGCUCUGCUUGCGGAAGCGGUAGAACGGAUUGUUGAUG ------.((((((((((-(-.((..(((.....))))).))).))-))).((((((((((.....))))))))((((((.((....))))))))..))......))). ( -42.90, z-score = -2.25, R) >droSim1.chr3R 10465979 99 - 27517382 ------GAUCGCGAGCG-G-ACUGAUGGAGCAUCCGAGCCCGCCU-UGCGCCGCAGCUCCUUCAAGGAGUUGCUCUGCUUGCGGAAGCGGUAGAACGGAUUGUUGAUG ------.((((((((((-(-.((..(((.....))))).))).))-))).((((((((((.....))))))))((((((.((....))))))))..))......))). ( -42.90, z-score = -2.16, R) >droSec1.super_0 3504689 99 - 21120651 ------GAUCGCGAGCG-G-ACUGAUGGAGCAUCCGAGCCCGCCU-UGCGCCGCAGCUCCUUCAAGGAGUUGCUCUGCUUGCGGAAGCGGUAGAACGGAUUGUUGAUG ------.((((((((((-(-.((..(((.....))))).))).))-))).((((((((((.....))))))))((((((.((....))))))))..))......))). ( -42.90, z-score = -2.16, R) >droYak2.chr3R 8292168 99 + 28832112 ------GAUCGAGAGCG-G-AUCGAGGGUGCAUCCGCUCCACCCU-UGCGCCGCAGCUCCUUCAAGGAGUUGCUUUGCUUGCGGAAGCGGUAGAACGGAUUGUUGAUG ------.((((((((((-(-..((((((((.........))))))-))..))((((((((.....))))))))...))))((....))..............))))). ( -40.20, z-score = -1.76, R) >droEre2.scaffold_4770 390411 99 - 17746568 ------GAUCGAGAGCG-G-ACUGAGGGAGCAUCCGCUCCACCCU-GGCGCCGCAGCUCCCUCAAGGAGUUGCUCUGCUUGCGGAAGCGGUAGAACGGAUUGUUGAUG ------.((((((((((-(-(.((......)))))))))((..((-(.....((((((((.....))))))))((((((.((....)))))))).)))..))))))). ( -41.60, z-score = -1.57, R) >droAna3.scaffold_13340 19517250 99 - 23697760 ------GAGCGGGAGCG-GGACCGGUGGGGUG-CCGGCCUACGCU-GGCGGUGGAGCUCUUUGAGGGAGUUGCUCUGCUUGCGGAAGCGGUAGAACGGAUUAUUAAUG ------..((....))(-((.(((((.....)-))))))).(((.-(((((.(.((((((.....)))))).).))))).)))....((......))........... ( -37.90, z-score = -1.14, R) >dp4.chr2 23178220 108 + 30794189 GACCGGGAGCGGGAGCGUGUGCCCUGGGCACGGUUGCGCCUCUUCAUGCGUCGCAGCUCCUUCAAGGAGUUACUCUGCUUGCGAAAGCGAUAGAACGGAUUGUUAAUG ..(((.(((((((.(((((((((...))))))...((((........)))))))((((((.....))))))..)))))))((....)).......))).......... ( -39.20, z-score = -0.40, R) >droPer1.super_3 5950606 108 + 7375914 GACCGGGAGCGAGAGCGUGUGCCCUGGGCACGGUUGCGCCUCUUCAGGCGUCGCAGCUCCUUCAAGGAGUUACUCUGCUUGCGAAAGCGAUAGAACGGAUUGUUAAUG ..(((.(((((((.(((((((((...))))))...((((((....)))))))))((((((.....)))))).))).))))((....)).......))).......... ( -43.20, z-score = -1.51, R) >droWil1.scaffold_181089 779102 96 + 12369635 ------GAUCUGCAACG-UAUACGUCGUGUUU-----GCCGACGCAAACGUCGCAGUUCUUUCAGCGAGUUGCUUUGCUUACGAAAGCGAUAGAACGGAUUGUUGAUA ------((.((((.(((-(...(((((.(...-----.))))))...)))).)))).))..((((((((((.((((((((....)))))).)))))...))))))).. ( -30.50, z-score = -1.22, R) >droVir3.scaffold_13047 7337518 102 + 19223366 ------GAUCUGUGGCGACGCAGCUGGCGACGCGCGCGUUUACGCAUGCGCCGCAGCUCCCUGAGGGAGUUGCUCUGCUUGCGAAAGCGAUAGAACGGAUUGUUGAUG ------((((((((((..(((.....)))..(((.(((....))).))))))(((((((((...)))))))))((((.((((....)))))))))))))))....... ( -47.70, z-score = -2.92, R) >droMoj3.scaffold_6540 3254449 90 + 34148556 ------GAUCUAUGCCGACGCAUCUGGCGAUG------------CAGGCGGCGCAGCUCGCGCAGCGAAUUGCUCUGCUUGCGAAAGCGGUAGAACGGAUUGUUGAUG ------(((((.(((((.(((.....))).((------------(((((((.((((.((((...)))).)))).)))))))))....)))))....)))))....... ( -36.60, z-score = -1.03, R) >droGri2.scaffold_14830 3179265 96 + 6267026 ------GAUCUGUGACGACGCAAUUGGCGACG---ACGU---CGCAAACGUCGCAACUCCCUCAAUGAGUUGCUCUGCUUGCGAAAGCGAUAAAAAGGAUUAUUGAUG ------....(((((((.(((.....)))...---.)))---))))..(((((((((((.......))))))).....((((....))))..............)))) ( -29.90, z-score = -2.10, R) >anoGam1.chr2R 46320426 92 + 62725911 -GGCUGGCCGAACCGAGCGUACGGCUUGCACUGUCCAGGCUGGCCAAACGCUGCCGGUGGGCCGACGGGCGGAUGUUUUCGUAAUUGUUGCGG--------------- -.((.((((..(((((((((..((((.((.((....)))).))))..)))))..)))).))))((((.(((((....)))))...))))))..--------------- ( -38.00, z-score = -0.16, R) >consensus ______GAUCGAGAGCG_G_ACCGUGGGGACGUCCGCGCCUCCCCAUGCGCCGCAGCUCCUUCAAGGAGUUGCUCUGCUUGCGAAAGCGGUAGAACGGAUUGUUGAUG .................................................(((((((((((.....))))))((.......))....)))))................. (-11.39 = -11.25 + -0.14)



| Location | 4,213,667 – 4,213,766 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 64.14 |
| Shannon entropy | 0.80512 |
| G+C content | 0.58541 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -8.79 |
| Energy contribution | -9.28 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4213667 99 - 27905053 CAUCAACAAUCCGUUCUACCGCUUCCGCAAGCAGAGCAACUCCUUGAAGGAGCUGCGGCGCA-AGACGGGCUCGGAUGCUCCAUCAGU-C-CGCUCGCGAUC------ ..................((((((....))))...(((.((((.....)))).)))))(((.-((.((((((..((((...)))))))-)-)))).)))...------ ( -30.80, z-score = -0.31, R) >droSim1.chr3R 10465979 99 + 27517382 CAUCAACAAUCCGUUCUACCGCUUCCGCAAGCAGAGCAACUCCUUGAAGGAGCUGCGGCGCA-AGGCGGGCUCGGAUGCUCCAUCAGU-C-CGCUCGCGAUC------ ..................((((((....))))...(((.((((.....)))).)))))(((.-.((((((((..((((...)))))))-)-)))).)))...------ ( -32.60, z-score = -0.36, R) >droSec1.super_0 3504689 99 + 21120651 CAUCAACAAUCCGUUCUACCGCUUCCGCAAGCAGAGCAACUCCUUGAAGGAGCUGCGGCGCA-AGGCGGGCUCGGAUGCUCCAUCAGU-C-CGCUCGCGAUC------ ..................((((((....))))...(((.((((.....)))).)))))(((.-.((((((((..((((...)))))))-)-)))).)))...------ ( -32.60, z-score = -0.36, R) >droYak2.chr3R 8292168 99 - 28832112 CAUCAACAAUCCGUUCUACCGCUUCCGCAAGCAAAGCAACUCCUUGAAGGAGCUGCGGCGCA-AGGGUGGAGCGGAUGCACCCUCGAU-C-CGCUCUCGAUC------ ........((((((((((((.(((.(((.......(((.((((.....)))).))).))).)-))))))))))))))......((((.-.-.....))))..------ ( -38.00, z-score = -2.67, R) >droEre2.scaffold_4770 390411 99 + 17746568 CAUCAACAAUCCGUUCUACCGCUUCCGCAAGCAGAGCAACUCCUUGAGGGAGCUGCGGCGCC-AGGGUGGAGCGGAUGCUCCCUCAGU-C-CGCUCUCGAUC------ .(((....((((((((((((((....))..((...(((.((((.....)))).))).))...-..))))))))))))((.........-.-.))....))).------ ( -35.50, z-score = -0.69, R) >droAna3.scaffold_13340 19517250 99 + 23697760 CAUUAAUAAUCCGUUCUACCGCUUCCGCAAGCAGAGCAACUCCCUCAAAGAGCUCCACCGCC-AGCGUAGGCCGG-CACCCCACCGGUCC-CGCUCCCGCUC------ ............(((((...((((....)))).(((.......)))..))))).........-((((..(((.((-.(((.....)))))-.)))..)))).------ ( -24.30, z-score = -1.90, R) >dp4.chr2 23178220 108 - 30794189 CAUUAACAAUCCGUUCUAUCGCUUUCGCAAGCAGAGUAACUCCUUGAAGGAGCUGCGACGCAUGAAGAGGCGCAACCGUGCCCAGGGCACACGCUCCCGCUCCCGGUC ...................((((((..((.((...(((.((((.....)))).)))...)).))..))))))..((((((((...)))))..((....))....))). ( -30.60, z-score = 0.17, R) >droPer1.super_3 5950606 108 - 7375914 CAUUAACAAUCCGUUCUAUCGCUUUCGCAAGCAGAGUAACUCCUUGAAGGAGCUGCGACGCCUGAAGAGGCGCAACCGUGCCCAGGGCACACGCUCUCGCUCCCGGUC ..........(((.......((((((....).)))))...........(((((.(((.(((((....))))).....(((((...))))).)))....)))))))).. ( -34.80, z-score = -0.88, R) >droWil1.scaffold_181089 779102 96 - 12369635 UAUCAACAAUCCGUUCUAUCGCUUUCGUAAGCAAAGCAACUCGCUGAAAGAACUGCGACGUUUGCGUCGGC-----AAACACGACGUAUA-CGUUGCAGAUC------ .(((........(((((.((((((....))))..(((.....))))).)))))((((((((.(((((((..-----.....))))))).)-)))))))))).------ ( -36.30, z-score = -4.73, R) >droVir3.scaffold_13047 7337518 102 - 19223366 CAUCAACAAUCCGUUCUAUCGCUUUCGCAAGCAGAGCAACUCCCUCAGGGAGCUGCGGCGCAUGCGUAAACGCGCGCGUCGCCAGCUGCGUCGCCACAGAUC------ ..............(((...((...((((.((...(((.(((((...))))).)))((((.((((((......)))))))))).))))))..))...)))..------ ( -38.20, z-score = -2.03, R) >droMoj3.scaffold_6540 3254449 90 - 34148556 CAUCAACAAUCCGUUCUACCGCUUUCGCAAGCAGAGCAAUUCGCUGCGCGAGCUGCGCCGCCUG------------CAUCGCCAGAUGCGUCGGCAUAGAUC------ ..............((((..((((((....).))))).....((.((....)).))((((..((------------((((....)))))).)))).))))..------ ( -31.60, z-score = -1.32, R) >droGri2.scaffold_14830 3179265 96 - 6267026 CAUCAAUAAUCCUUUUUAUCGCUUUCGCAAGCAGAGCAACUCAUUGAGGGAGUUGCGACGUUUGCG---ACGU---CGUCGCCAAUUGCGUCGUCACAGAUC------ .(((.....((((((.....((((((....).)))))........))))))((.(((((((..(((---((..---.))))).....))))))).)).))).------ ( -30.12, z-score = -2.19, R) >anoGam1.chr2R 46320426 92 - 62725911 ---------------CCGCAACAAUUACGAAAACAUCCGCCCGUCGGCCCACCGGCAGCGUUUGGCCAGCCUGGACAGUGCAAGCCGUACGCUCGGUUCGGCCAGCC- ---------------..((..................(((..(((((....))))).)))..(((((((((.((...((((.....)))).)).)))).))))))).- ( -31.40, z-score = -0.33, R) >consensus CAUCAACAAUCCGUUCUACCGCUUUCGCAAGCAGAGCAACUCCUUGAAGGAGCUGCGGCGCAUGGGGAGGCGCGGACGCCCCCACAGU_C_CGCUCCCGAUC______ ....................((((....))))...(((.((((.....)))).))).................................................... ( -8.79 = -9.28 + 0.49)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:14 2011