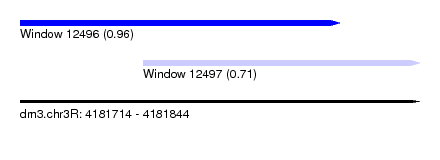
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,181,714 – 4,181,844 |
| Length | 130 |
| Max. P | 0.955509 |

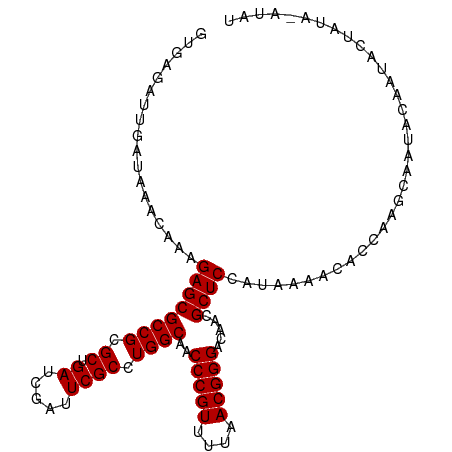
| Location | 4,181,714 – 4,181,818 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Shannon entropy | 0.13854 |
| G+C content | 0.45870 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4181714 104 + 27905053 GUGAGAUUGAUAAACAAAGAGCGCCGCGCUGAUCGAUUCGCUUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAGCCAAUACAAAACUAUA-CUAU (((...(((.....))).((((((((.((.((.....)))).))))..(((((....))))).....)))).......)))...................-.... ( -23.90, z-score = -2.16, R) >droSim1.chr3R 10434348 104 - 27517382 GUGAGAUUGAUAAACAAAGAGCGCCGCGCUGAUCGAUUCGCCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAAGCGAUACAAUCCUAUA-AUAU (((.(((((.........((((((((.((.((.....)))).))))..(((((....))))).....))))..........(....)...))))).))).-.... ( -25.00, z-score = -2.02, R) >droSec1.super_0 3473101 99 - 21120651 GUGAGAUUGAUAAACAAAGAGCGCCGCGCUGAUCGAUUCGCCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAAGCGAUACAAUACUAU------ (((...(((.....))).((((((((.((.((.....)))).))))..(((((....))))).....)))).......)))..................------ ( -23.90, z-score = -1.81, R) >droEre2.scaffold_4770 358048 103 - 17746568 GUGCGAUUGAUAAACAAAGAGCGCCGCGCUGAUCGAUUCGCCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAAC--CAAGCAAUACUAUACUAUUUAUAU .(((..(((.....))).((((((((.((.((.....)))).))))..(((((....))))).....))))........--...))).................. ( -25.10, z-score = -1.98, R) >consensus GUGAGAUUGAUAAACAAAGAGCGCCGCGCUGAUCGAUUCGCCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAAGCAAUACAAUACUAUA_AUAU ..................((((((((.((.((.....)))).))))..(((((....))))).....)))).................................. (-22.70 = -22.70 + -0.00)

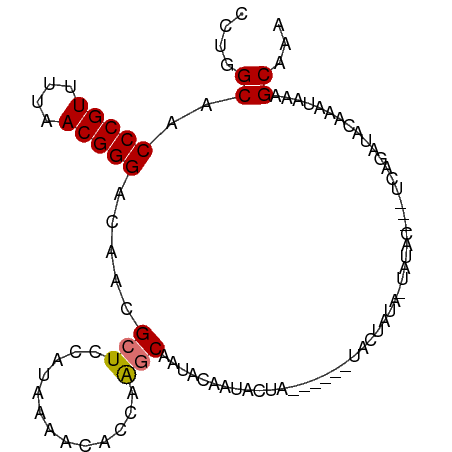
| Location | 4,181,754 – 4,181,844 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.26 |
| Shannon entropy | 0.22848 |
| G+C content | 0.37268 |
| Mean single sequence MFE | -13.29 |
| Consensus MFE | -9.38 |
| Energy contribution | -9.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4181754 90 + 27905053 CUUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAGCCAAUACAAAACUA------UACUAUA-UAUAC---UCAGAUACAAAUAAAGCAAA .(((((.....((((((...(((......))).))))))....)))))..........------.......-.....---.................... ( -11.40, z-score = -1.56, R) >droSim1.chr3R 10434388 90 - 27517382 CCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAAGCGAUACAAUCCUA------UAAUAUA-UAUAC---UCAGAUACAAAUAAAGCACA .((((...(((((....)))))....((((.............))))...........------.......-.....---))))................ ( -13.02, z-score = -1.78, R) >droSec1.super_0 3473141 85 - 21120651 CCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAAGCGAUACAA-----------UACUAUA-UAUAC---UCAGAUACAAAUAAAGCAAA .((((...(((((....)))))....((((.............))))......-----------.......-.....---))))................ ( -13.02, z-score = -2.02, R) >droEre2.scaffold_4770 358088 98 - 17746568 CCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAAC--CAAGCAAUACUAUACUAUUUAUAUACUAUACUAUAUGAAUCAGAUACAAAAAAAGCACA ....((..(((((....))))).....(((.........--..))).........(((((((((((......))))))))).))...........))... ( -15.70, z-score = -2.72, R) >consensus CCUGGCAACCCGUUUUAACGGGACAACGCUCCAUAAAACACCAAGCAAUACAAUACUA______UACUAUA_UAUAC___UCAGAUACAAAUAAAGCAAA ....((..(((((....))))).....(((.............))).................................................))... ( -9.38 = -9.45 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:12 2011